The figure summarizes the whole idea at the end of the project design. You can find all details about the different parts of the project below.
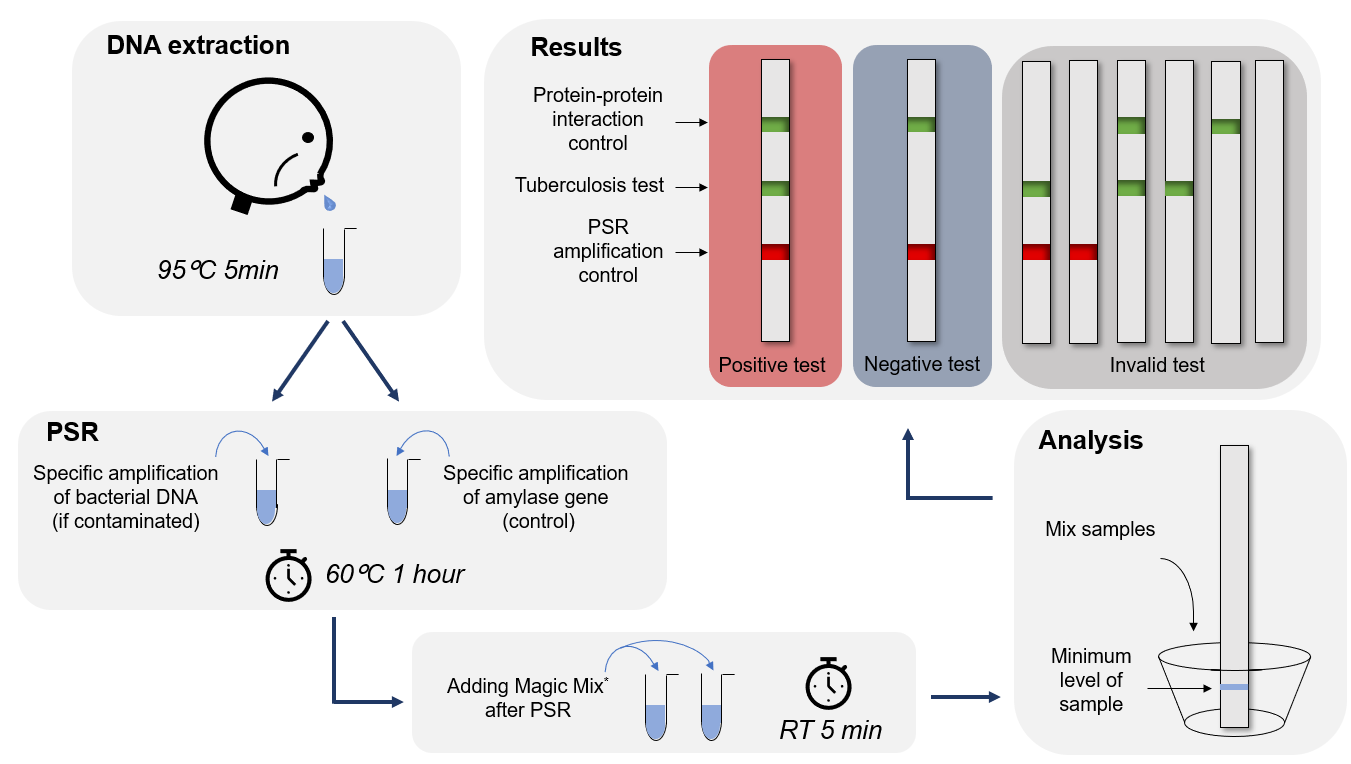
PSR stands for polymerase spiral reaction. It is a novel isothermal nucleic acid amplification method that only requires one pair of primers and one enzyme like PCR (Polymerase Chain Reaction) with high specificity, efficiency, and rapidity under isothermal conditions.
The PSR is performed at constant temperature (61–65 °C). Betaine unlocks the double-stranded DNA. This reaction could be done within an hour with a high sensitivity and specificity to detect the specific target. The PSR takes advantage of different methods : only one set of primers is needed (same as the PCR) and amplification is done at constant temperature such as the LAMP(Loop-mediated isothermal amplification) assay.
Description
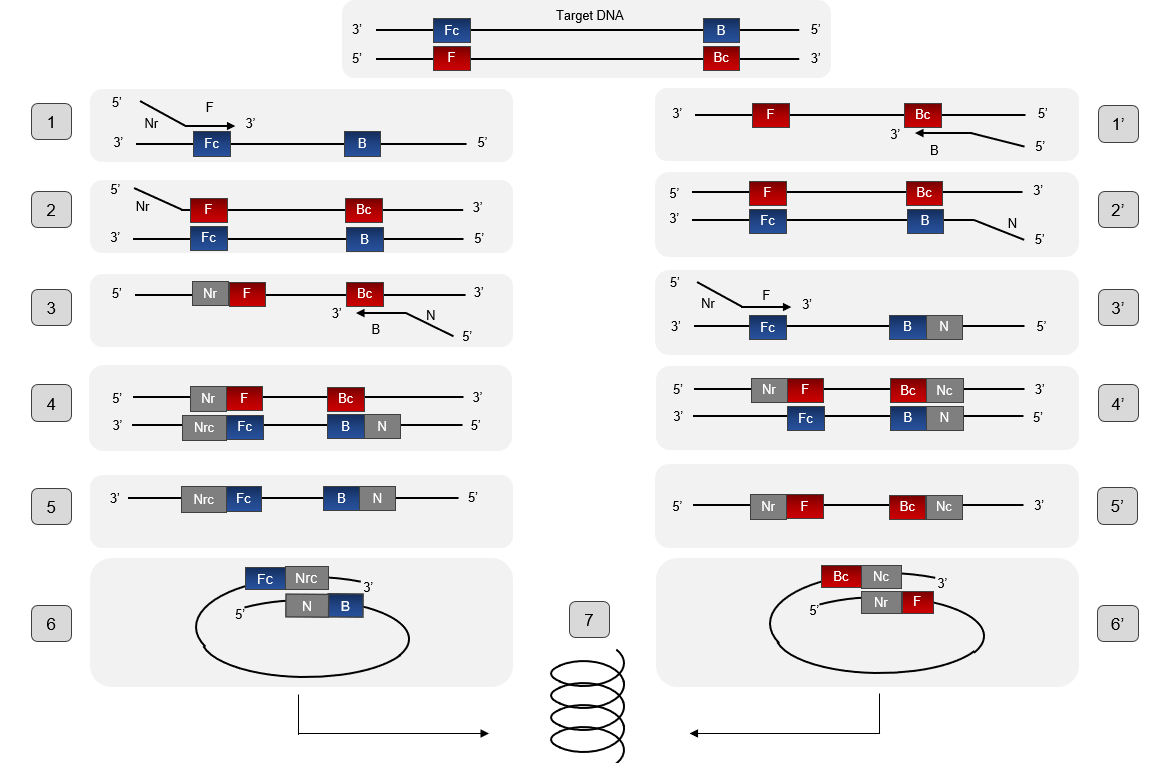
The mechanism of the PSR process is illustrated in Figure. To understand the mechanism of PSR, it is important to know beforehand the particular structure of the pair of primers. Each primer is composed of a complementary part to the target sequence, here F stand for the primer Ft or B stand for the primer Bt, and they are as well a reverse part and complementary to each other, and Nr stand for the primer Ft or B stand for the primer Bt. Betain is an agent that allows the departure of the PSR reaction. Its function is to reduce secondary structure formation when the DNA matrix is rich in GC. In addition, it helps Bst polymerase which has strand displacement activity to separate double strands of DNA when the temperature reaches at 60° C. When the double-stranded structure unlocks, the complementary part to the target sequence of the primer Ft just hybridize and then will extend thanks to the 5'-3 'polymerase activity of the Bst polymerase (structure 1 and 2), thus forming the complementary strand. Then, the primer Bt will hybridize in a complementary manner on the newformed strand and also extend (structure 3 and 4). So, at this stage we have formed a strand that has the sequences Nr and N which are inverse and complementary to it (structure 5) which will wrap on itself to form a loop (structure 6). The 3 'end of Nrc will continue to extend on itself to finally form a kind of spiral (structure 7). The extension mechanism of the second strand follows the same principle. PSR assay can be monitored continuously in a real-time turbid meter instrument or visually detected using fluorescent dye (SYBR Green I) or metal ion indicator dyes (HNB and Calcein) and other detection methods used for LAMP method (1).
Our Amplification Molecular Diagnostics Project uses Advanced Polymerase Technology targeting the katG and rpoB genes. These two genes are responsible for 98% of the resistance to rifampicin and isoniazid. A genetic mutation at the position 315 of the gene katG leads to a partial loss of catalase-peroxidase activity and thus to the resistance to Isoniazid. The gene rpoB encodes for the beta subunit of the RNA polymerase and mutations are predominantly at codons 531, 526, and 516.
This technique is characterized by its speed and robustness. Our test is suitable for peripheral laboratories, due to the minimum material required.
These are the advantages of this diagnostic tool:
- Quick access to results
- High sensitivity and specificity
- Consolidated and virtually maintenance-free platform
- Robustness with inhibitors and reaction conditions that may have a negative impact on PCR methods.
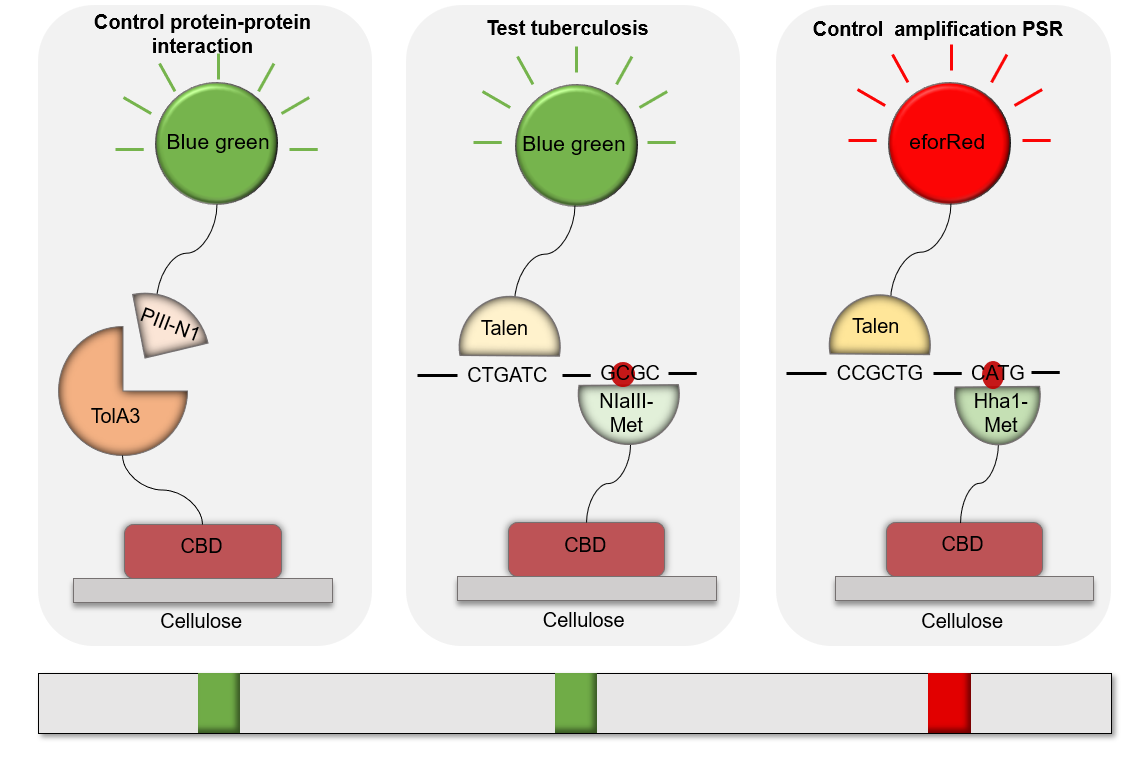
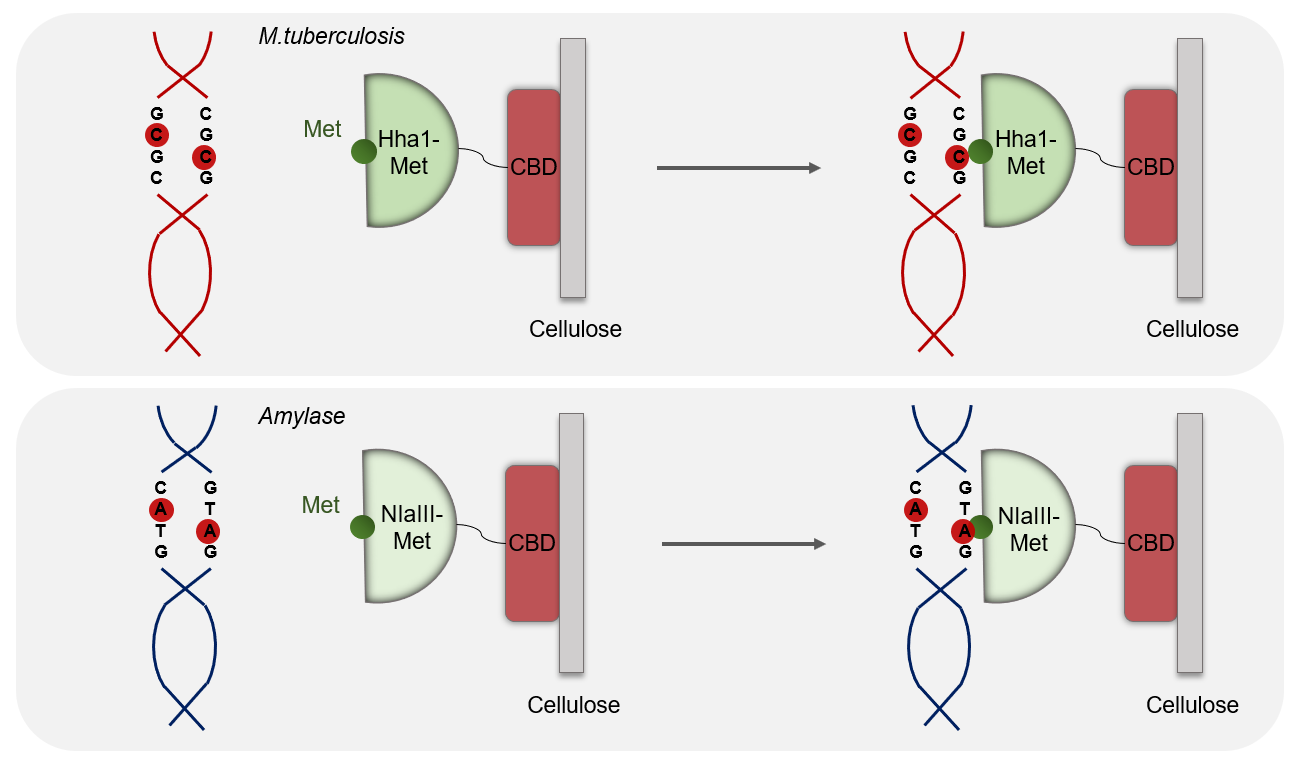
Methylase are enzyme that recognize a specific region of the DNA without cleaving it. We chose two methylases specific for each DNA. The aim of our process was to specifically capture Mycobacterium tuberculosis and amylase's DNA without cleaving or damaging them. We've been first thinking about the Streptavidine/Biotine system. After several meetings and discussions, we found out which methylase would match with our project. We chose HhaI-M for tuberculosis that comes from the Haemophilus parahaemolyticus organism that recognizes the GCGC sequence and that methylates C-2 on both strands and thus protects the latter from cleavage.
HhaI-M sequence
MIEIKDKQLTGLRFIDLFAGLGGFRLALESCGAECVYSNEWDKYAQEVYEMNFGEKPEGD ITQVNEKTIPDHDILCAGFPCQAFSISGKQKGFEDSRGTLFFDIARIVREKKPKVVFMEN VKNFASHDNGNTLEVVKNTMNELDYSFHAKVLNALDYGIPQKRERIYMICFRNDLNIQNF QFPKPFELNTFVKDLLLPDSEVEHLVIDRKDLVMTNQEIEQTTPKTVRLGIVGKGGQGER IYSTRGIAITLSAYGGGIFAKTGGYLVNGKTRKLHPRECARVMGYPDSYKVHPSTSQAYK QFGNSVVINVLQYIAYNIGSSLNFKPY
For the amylase we've decided to choose NlaIII-M which comes from Neisseria lactamica and which recognizes the CATG sequence and methylates A-2 on both strands to protect the latter from cleavage.
NlaIII-M sequence
MNYIGSKLKLSNWLETEISNVAGHSLSDKVFCDLFAGTGIVGRKFKTNVKQVIANDMEYY SYVLNRNYIGNCQSILKAGELLQRLEQLPPREGLIYQHYCLGSGSERQYFSDENGKKIDA VRIQIEEWKNTRYIDEDTYYFLLATLLEGADKVANTASVYGAFLKNLKKSALKPLSLEPA LFEIGSDGHQVYQADANQLIKNISGDILYLDPPYNARQYGANYHLLNSIALYDDFTPKGK TGLREYSRSKYCSKSDVVPVFEALIRDADFQYIFLSYNNEGLMSVGQVREIFERFGKYDL VQTEYRRFKADKTENRNHKANSTFEYLHILEKTF
These two enzymes are specific for each DNA because they are not found in the two DNAs. So, we increase the specificity of our test thanks to the methylases.
Why we decided to choose these proteins ?
NlaIII-mehtylase and HhaI-methylase are two proteins that have high affinity and generate strong protein-DNA interactions. Using these proteins for the best would be the best option for us to detect the targeted DNA such as the human amylase gene, katG and rpoB present in Mycobacterium tuberculosis genome. So after amplification, these two gene would be recognized by the methylase and create DNA/protein complex.
In our project both proteins will be fused with a cellulose binding domain in order to enable the binding of NlaIII and HhaI proteins on a cellulose membrane. This membrane is the main support of our kit. Both fusions are made on the C-terminal domains.
About the Design
We've add a 6Hig-Tag and a TeV cleavage site into the NlaIII-CBD and HhaI sequences in order to facilitate the purification. The codons have been optimized in order to match with protein expression into the K12 E.coli. To perform the production of NlaIII-CBD and HhaI proteins in a K12 E.coli strain, T7 promotor and a RBS, that are essential regulatore for the transcription, were added in the pSB1C3.
This biobrick is in RFC 10 standards with: The prefixe (with ATG in frame): 5' GAATTC GCGGCCGC T TCTAGA TG GCCGGC and the suffixe (contains Stop in frame): ACCGGT TAAT ACTAGT A GCGGCCG CTGCAG 3'
NlaIII-Methyase construction

HhaI-Methyase construction

What is a TALs effector
It’s a transcription activator-like (TALs) effectors that is a newly described class of specific DNA binding protein. This molecule is produced by a pathogenic bacteria of the genus Xanthomonas, via their type III secretion system. These proteins are able to bind to promoter sequences in the host plant and to activate the expression of genes involved in bacterial infection.
How do TALs function
TALs effectors have attracted great interest as DNA targeting tools. Their specificity is determined by a central tandem domain, 33–35 amino acid repeats, followed by a single truncated repeat of 20 amino acids. Most of the TAL effectors are characterized by 12 to 27 full repeats. A polymorphic pair of adjacent residues at positions 12 and 13 in each repeat, the ‘repeat-variable di-residue’ (RVD), specifies the target, one RVD to one nucleotide, with the four most common RVDs each preferentially associating with one of the four bases (1).
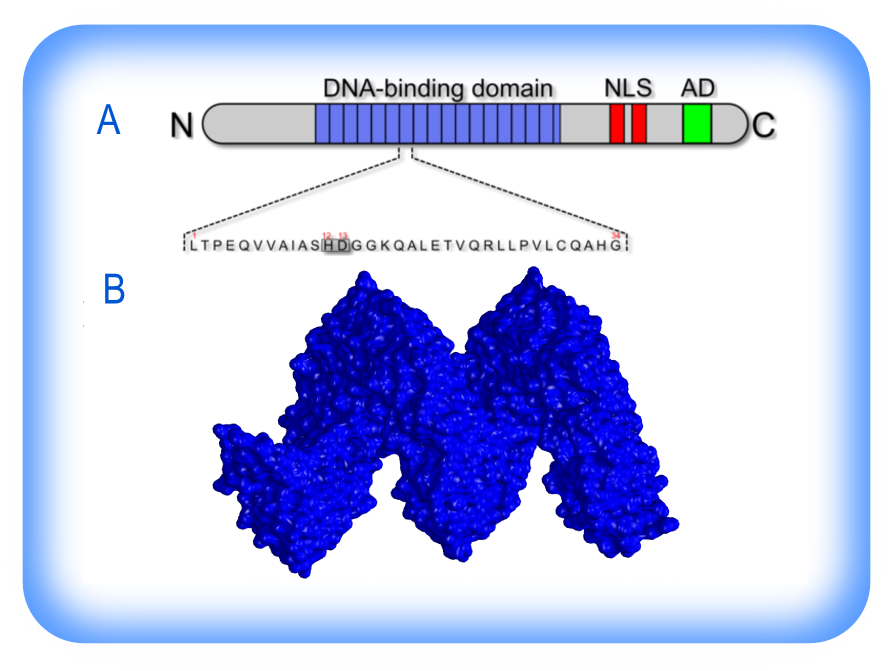
The structure of TALs effectors. (A) Schematic representation of TALs effector structure and its DNA-binding domain (bleu), containing multiple 34 amino acids tandem repeats with RVDs at the 12th and 13th residue (Scholze and Boch, 2011). (B) 3D structure of a TALs effector.
Why did we choose TALs ?
TALs technology is currently revolutionizing synthetic biology, not only because of higher sequence fidelity or less cytotoxicity compared to other DNA binding proteins. Mainly because it can bind known DNA sequences, whereas zinc fingers proteins have to be selected post-library screening.
CRISPR/Cas9 is composed of a non-specific Cas9 nuclease and a set of programmable sequence-specific, CRISPR RNA guide (crRNA). The complex might not be stable in any environment, and can guide Cas9 to cleave DNA and generate double-strand breaks at target sites, due to the non-specific interaction between enzyme-fused Cas9 proteins and gRNA. This phenomenon makes the system unpredictable and uncontrollable (3). That’s why TALs technology is generally much cheaper, less time consuming and does guarantee binding sites for every predefined sequences. As mentioned before, the key of developing a successful in vivo scaffold system is to establish highly-specific and efficient interaction with DNA fragments.
How did we assembly our TALs ?
We've based our strategy on an article from Nucleic Acids Research called “Efficient design and assembly of custom TALs and other TAL effector-based constructs for DNA targeting”. In addition to this article, we 've found complementary sequences, that allowed us to construct our TALs. The designed TALs share a similar structure and are able to bind to any DNA sequence of interest with high-specificity and efficiency.
What is the role of TALs in this project ?
We've designed TALs technology in such a way that they can be produced in prokaryotic organisms by optimizing the TALs coding sequence. Those TALs were designed and directly ordered from IDT and were transcriptional fusion with a chromoprotein on the C-terminal. This fusion is used to recognize a DNA fragment that were amplify by PSR. TALs-chromoprotein is used as a color signal when bound to the targeted fragment. Thus the amplicon would be recognized by the methylase/CBD complex and the TALs/chromprotein complex. The fixation of each fragment form a colored strip, which allows us to read the results of the test.
Designing of the scaffold and its corresponding engineered TALs effectors
In our project, we designed three different DNA Binding motifs (DNA BMs). The sequences were chosen from different DNA that while be amplify by PSR, the first one is from a human amylase gene (TAL1), the second one is able to recognize either Tuberculosis gene or Smegmatis gene that are non-resistant (TAL2). For the last one we were able to engineer a TAL that’s able to bind itself on to a drug-resistant mycobacterium tuberculosis of the strain UKR100 that codes for “16S ribosomal RNA gene”, that are mutated at the rpoB gene at three different codons 531, 526 and 516 (TAL3).
Those BMs are:
BM1: 5’ CTGATC 3’ (in Amylase fragment)
BM2: 5’ CCGCTG 3’ (in Tuberculosis or Smegmatis non-resistant fragment)
BM3: 5’ CTGTCG 3’ (in Tuberculosis UKR100 strain resistant fragment)
Based on the sequences we chosen, we engineered three different TAL effectors. TAL1 can specifically recognize the sequence of BM1, TAL2 can specifically recognize the sequence of BM2 and TAL3 can specifically recognize the sequence of BM3.
Sequence of TAL1
MHHHHHHENLYFQGMPLNLTPDQVVAIASHDGGKQALETVQRLLPVLCQDHGLTPDQVVA IASNGGGKQALETVQRLLPVLCQDHGLTPDQVVAIASNNGGKQALETVQRLLPVLCQDHG LTPDQVVAIASNIGGKQALETVQRLLPVLCQDHGLTPDQVVAIASNGGGKQALETVQGGK QALETVQRLLPVLCQDHGAIASHDGGKQALESIVAQLSRPDPALAALTN
Sequence of TAL2
MHHHHHHENLYFQGMPLNLTPDQVVAIASHDGGKQALETVQRLLPVLCQDHGLTPDQVVA IASHDGGKQALETVQRLLPVLCQDHGLTPDQVVAIASNNGGKQALETVQRLLPVLCQDHG LTPDQVVAIASHDGGKQALETVQRLLPVLCQDHGLTPDQVVAIASNGGGKQALETVQGGK QALETVQRLLPVLCQDHGAIASNKGGKQALESIVAQLSRPDPALAALTN
Sequence of TAL3
MHHHHHHENLYFQGMPLNLTPDQVVAIASHDGGKQALETVQRLLPVLCQDHGLTPDQVVA IASNGGGKQALETVQRLLPVLCQDHGLTPDQVVAIASNNGGKQALETVQRLLPVLCQDHG LTPDQVVAIASNGGGKQALETVQRLLPVLCQDHGLTPDQVVAIASHDGGKQALETVQGGK QALETVQRLLPVLCQDHGAIASNNGGKQALESIVAQLSRPDPALAALTN
Prototypes
We received our assembly TALs from IDT without the TAL N-terminal, TAL C-terminal, attL1 or attL2 homologous arms, because we were enabling all possibilities of our fragment being cut by the TALES. Otherwise, it’s missing a promoter and T7 RBS so that our prokaryotic bacteria are able to produce our proteins. We started by using two primers that we designed to amplify our fragment by PCR, that contains EcoRI, XbaI has a forward primer and SpeI, PstI as a reverse primer, that are restriction enzyme sites present on either of our sequence. The digestion and ligation strategy was used to ligate our fragment in a vector behind a promoter T7 RBS. BBa_K2915225 corresponds to TAL1 with its promoter T7 and RBS, BBa_K2915275 is TAL2 with its promoter T7 and RBS and BBa_K2915285 is TAL3 with its promoter T7 and RBS.
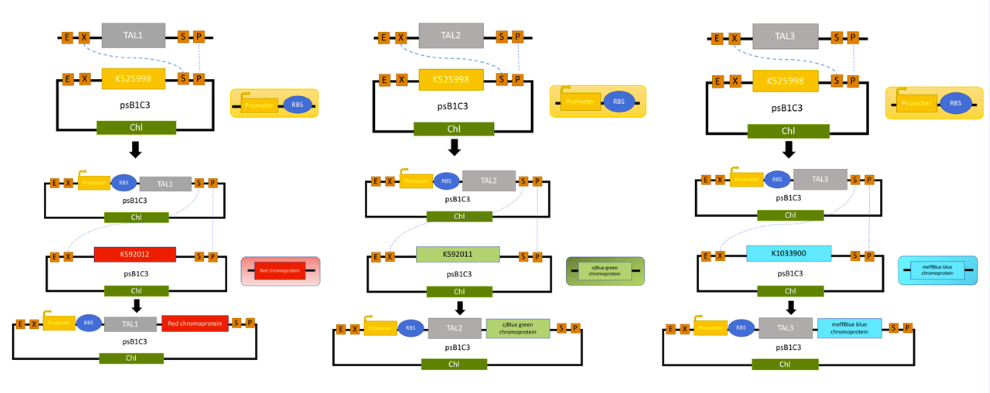
TAL1 was design to recognized an Amylase gene, which is use has a control in our test. On the 1st/red band means that the sample was from a human patient and/or that our amplification by PSR works correctly.
TAL2 was design to recognized the gene rpoB or katG gene from M. tuberculosis, and M. smegmatis, which is use has a positive band in our test. The 2nd/Green band is visible when the patient is infected with M. tuberculosis. TAL2 are also design to detect the non-resistant M. tuberculosis, because it was design to region that aren’t mutated.
TAL3 was design to improve our test by detecting only the rpoB gene from M. tuberculosis on one of its mutant (526 amino acid), which is use has a positive band in our test that are able to detect a drug-resistant M. tuberculosis. The 2nd band is blue when the sample amplify present that resistant strain. This TAL was design to integrate human practices, see more.
About the Design
We've added a 6Hig-Tag and a TeV cleavage site into the TAl1, TAL2 and TAL3 sequences to facilitate the purification. We've optimized the codons enabling the production into the K12 E. coli strain. To perform the production of TAl1, TAL2 and TAL3 protein in a K12 E. coli strain, we've added a T7 promotor and a RBS into the pSB1C3.
This biobrick is in RFC 10 standards with: The prefixe (with ATG in frame): 5' GAATTC GCGGCCGC T TCTAGA TG GCCGGC and the suffixe (contain Stop in frame): ACCGGT TAAT ACTAGT A GCGGCCG CTGCAG 3'
TAL1 construction in the pSB1C3 plasmid

TAL2 construction in the pSB1C3 plasmid

Reference
- Scholze, H., and Boch, J. (2011) TAL effectors are remote controls for gene activation. Curr. Opin. Microbiol. 14, 47-53.
- Maeder, M. L. et al. Rapid ‘Open-Source’ Engineering of Customized Zinc-Finger Nucleases for Highly Efficient Gene Modification. Molecular Cell 31, 294–301 (2008).
- Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges . Human Molecular Genetics, 2014, 23(R1):R40.
- Cermak T, et al. Efficient design and assembly of custom TALEs and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 2011 Sep 1;39(17):7879.
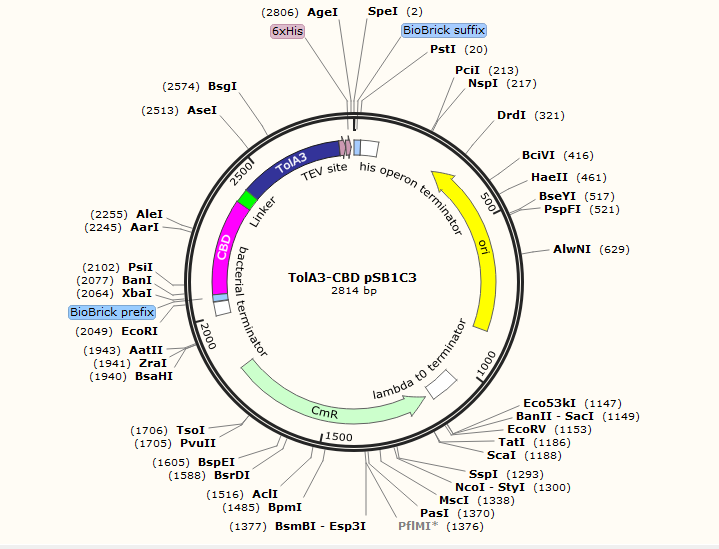
TolA protein
Structure
TolA is part of the Tol/Pal system which is involved in cell division and membrane integrity. It’s located in the inner membrane and is composed of three domains: the N-terminal Domain I (TolAI), from residues 1-47 including a 20-residues hydrophobic membrane, a spanning region that anchors the protein to the cytoplasmic membrane; Domain II (TolAII), from residues 48-301, which forms a rigid helix connecting the domains on both side of it; and the C-terminal Domain III (TolAIII) from residues 302-421, which may be involved in the function of TolA by interacting with the periplasmic or outer membrane proteins, due to the binding of domain II. For additional details see Tol.
C-terminal Domain
The C-terminal domain of TolA is directly involved with the N-terminal of both Colicin and the phage minor coat gene 3 protein.
Function
TolA could act as a bridging protein, connecting the inner and outer membranes. TolA is involved in import mechanism such as the uptake of bacteriotoxins (Colicin), and filamentous bacteriophage’s DNA. This protein might be an actor of the bacterial sensitivity to these groups, in particular the alpha helix of domain II. A deletion in this region raises cellular sensitivity to deoxycholate.
Sequence
GGGAKGNNASPAGSGNTKNNGASGADINNYAGQIKSAIESKFYDASSYAGKTCTLRIKLA PDGMLLDIKPEGGDPALCQAALAAAKLAKIPKPPSQAVYEVFKNAPLDFKP
g3P protein
Structure
The minor P3 coat protein located at the distal end of the phage has between three and five copies. The two proteins are composed of three distinct domains separated by two regions of low complexity that serve as linkers. Although the N-terminal domains (N1) and the central domains (N2) are exposed on the surface of the capsid, the C-terminal domain (N3) anchors the P3 protein to the phage particle through hydrophobic interactions.
N1-Domain
TheP3- N1 domain of P3 protein is directly involved with the TolA3 C-terminal .
Sequence
MKKLLFAIPLVVPFYSHSAETVESCLAKPHTENSFTNVWKDDKTLDRYAN YEGCLWNATGVVVCTGDETQCYGTWVPIGLAIPENEGGGSEGGGSEGGGS EGG
References
- Laetitia Houot et al, Electrostatic interactions between the CTX phage minor coat protein and the bacterial host receptor TolA drive the pathogenic conversion ofVibrio cholerae, DOI 10.1074/jbc.M117.786061
- Andrew J. Heilpern and Andmatthew K. Waldor, CTX Infection of Vibrio cholerae Requires the tolQRA Gene Products, Journal of Bacteriology, Mar. 2000, p. 1739–1747
Why those proteins are in part of our project
P3-N1 M13 and TolAEC are two proteins that have high affinity and generate strong protein-protein interactions. The use of these two proteins as internal control of this method would make it possible to verify that the interactions realized during the test are not ramdom interactions and would also allow our test to verify that the produced chromoproteins is functional and clearly visible.
In our project both proteins will be fused to two other protein domains. Domain 3 of the TolA protein was fused to a cellulose binding domain in order to allow the attachment of the TolA protein on a cellulose membrane wich is a support for our kit.
The P3 protein have been fused to a green chromoprotein thus allowing the P3-N1 / TolA3 complex to be visualized on the cellulose membrane. The presence of the complex will be visible by the appearance of a green band on the strip test.
About the design in the pSB1C3 plasmid
We add a 6Hig-Tag and a TeV cleavage site into the TolA3-CBD and P3-N1-Green chromoprotein sequences which allows us to purify our proteins. We optimized the codons to be able to produce these proteins into the K12 E.coli strain. To perform the production of TolA3-CBD and the P3-N1-Green chromoprotein proteins in a K12 E.coli strain, we added a T7 promotor and a RBS into the pSB1C3 plasmide,which is the regulator.
This biobrick is in RFC 10 standards with: The prefixe (with ATG in frame): 5' GAATTC GCGGCCGC T TCTAGA TG GCCGGC and the suffixe (contain Stop in frame): ACCGGT TAAT ACTAGT A GCGGCCG CTGCAG 3'

Antifreeze proteins (AFPs) protect certain cold-adapted organisms from freezing to death by selectively adsorbing to internal ice crystals and inhibiting ice propagation. The molecular details of AFP adsorption-inhibition is uncertain but is proposed to involve the Gibbs–Thomson effect. Here we show by using unbiased molecular dynamics simulations a protein structure-function mechanism for the spruce budworm Choristoneura fumiferana AFP, including stereo-specific binding and consequential melting and freezing inhibition. The protein binds indirectly to the prism ice face through a linear array of ordered water molecules that are structurally distinct from the ice. Mutation of the ice binding surface disrupts water-ordering and abolishes activity. The adsorption is virtually irreversible, and we confirm the ice growth inhibition is consistent with the Gibbs–Thomson law.
In our project, we'll test three kind of antifreeze proteins coming from different species (yeast, fish, butterfly), in order to determine which one of these proteins is the most efficient.
Choristoneura fumiferana (Spruce budworm moth) (Archips fumiferana) AFP-Iu1
Choristoneura fumiferana, the spruce budworm, has a freeze-avoidance strategy for overwintering, they can produce low molecular weight cryoprotectants to lower the freezing point of their hemolymph, avoid the initiation of ice growth by sealing themselves in cocoons, eliminate materials which could act as nucleators, increase production of energy sources, and even produce heat shock proteins (Hsps) and other stress proteins for protection against cold shock and injury, allow the larvae to survive temperatures of –30 °C or lower.
Similar to all AFPs from metazoans that have been characterized to date, AFPs in C. fumiferana are encoded by a multicopy gene family. For example, there are 30 – 40 copies of both liver and skin-type AFP genes in winter flounder (Scott et al., 1985; Gong et al., 1996). Unlike the case for the Type I AFP, where there is a 40% increase in levels of tRNA alanine isoacceptors in winter-caught fish (Pickett et al., 1983), there is no evidence of translational control of CfAFP expression; Western blots indicated that AFP was present in first instars, most abundant in second instars (independent of temperature) and declined after a week post diapause. Together, the Western and Northern analysis as well as the half-life determinations, show that the expression of CfAFP genes is not limited to the overwintering stage and that there is variation from gene to gene that cannot be attributed to 3′UTR-determined destabilizing elements.
Sequence:
MEYFFLLIVLAIRTVSSDGTCVNTNSQITANSQCVKSTATNCYIDNSQLV DTSICTRSQYSDANVKKSVTTDCNIDKSQVYLTTCTGSQYNGIYIRSSTT TGTSISGPGCSISTCTITRGVATPAAACKISGCSLSAM

sp. (strain AY30) (Arctic yeast)
Ice-binding proteins (IBPs) are a class of protein that has an affinity to ice. Some IBPs, for example fish antifreeze proteins (AFPs) are a biological antifreeze, which bind to the ice surface and subsequently inhibit further growth of the ice crystal. This behavior of IBP eventually lowers the freezing point of the solution, and hence creates a gap between freezing and melting points. This activity is called thermal hysteresis (TH).
Recently, we have identified two IBPs: one (designated as LeIBP) from the Arctic yeast Glaciozyma sp. and the other (designated as FfIBP) from the Antarctic sea ice bacterium, Flavobacterium frigoris. Two IBPs share 56% sequence identity and have almost the same β-helical structure. They belong to the same β-helical family of IBPs. Since both of the IBPs are produced by organisms overwintering in a brine channel and have N-terminal signal peptide, their main role seems to protect the organisms themselves from freezing damage. Both of the IBPs are known to possess TH and IRI activities. Despite their high similarity in primary and tertiary structure, LeIBP is moderately active (0.34 °C at 10.8 mg/mL) in TH, while FfIBP is hyperactive (2.2 °C at 0.13 mg/mL). This is because of the fact that FfIBP has more prominent ice-binding regular motifs (T-A/G-X-T/N motif) and ice-binding residues arrayed regularly on its ice-binding site than LeIBP. However, their IRIshowed opposite results. LeIBP exhibited IRI down to 0.001 mg/mL concentration (37 nM), while FfIBP did to 0.028 mg/mL (2.5 μM). This indicates that TH activity is not necessarily proportional to IRI. IRI activity of LeIBP is relatively high compared to fish and other β-helical IBPs. Recently, a few bacterial IBPs having almost identical β-helical structure to LeIBP and FfIBP were reported.
Sequence:
MSLLSIITIGLAGLGGLVNGQRDLSVELGVASNFAILAKAGISSVPDSAI LGDIGVSPAAATYITGFGLTQDSSTTYATSPQVTGLIYAADYSTPTPNYL AAAVANAETAYNQAAGFVDPDFLELGAGELRDQTLVPGLYKWTSSVSVPT DLTFEGNGDATWVFQIAGGLSLADGVAFTLAGGANSTNIAFQVGDDVTVG KGAHFEGVLLAKRFVTLQTGSSLNGRVLSQTEVALQKATVNSPFVPAPEV VQKRSNARQWL
Dendroides canadensis thaumatin-like protein 2
Antifreeze proteins (AFPs) decrease the non-equilibrium freezing point of water while not affecting the melting point. This results in a difference between the freezing and melting points that is called Thermal Hysteresis. According to the adsorption-inhibition mechanism of action, the AFPs decreases the freezing point by adsorbing onto the surface of ice crystals at specific growth sites, thanks to hydrogen bonding, hydrophobic, and/or van der Waals interactions. This adsorption enables ice crystal growth to occur only between the AFPs(in highly curved (high surface free energy) rather than the low- curved (low surface free energy)). Therefore, the temperature must be decreased below the equilibrium melting point before ice crystal growth can take place.
Overwintering larvae of the beetle Dendroides canadensis produce a family of antifreeze proteins (DAFPs) that function to inhibit inoculative freezing across the cuticle and to promote supercooling by inhibiting ice nucleators. The group I DAFPs are present in the hemolymph, while groups II and III DAFPs occur in the gut fluid. There are both low-molecular mass and high-molecular mass antifreeze protein enhancers present in the hemolymph, and these significantly increase the thermal hysteresis activity (THA). The protein enhancers interact with the DAFPs, forming complexes that, because they are larger than the DAFP alone, block a larger surface area of the
potential seed ice crystal than does the DAFP alone. Therefore, THA is increased. Because the protein enhancers are thought to bind to the DAFPs, a yeast two-hybrid screen was used to identify D. canadensis proteins that interact with DAFPs and might therefore be potential enhancers. This screen previously demonstrated that certain group I DAFPs found in the hemolymph interact with one another, and these were then shown to enhance the thermal hysteresis activities of one another. The study reported here demonstrates that a thaumatin-like protein (until recently known only from plants) was also identified by the yeast two-hybrid screen as interacting with DAFP-1 and -2. The recombinant thaumatin-like protein was subsequently shown to enhance DAFP-1 and -2 activities.
Sequence:
MFVLLVAALLATAQAVEFHLQNNEPGPVWVGIQGNPEHTHLSNGGLILDQ GQGVVLQAEDNWAGRFWGRTWCDPATNHCQTGDCGNKIECAGAGGTPPAS LAEITLKGWGDLDYYDISLVDGFNMRIAVSGFVVNFDSSLTFVSDSSNPL METEMVANTVANVVNALLISTTTALVN-

About the Design
We add a 6Hig-Tag and a TeV cleavage site into the AFP-lu1 sequence which allows us to purify our proteins. We optimized the codons to be able to produce these proteins into the K12 E. coli strain. To perform the production of AFP-lu1 protein in a K12 E. coli strain, we added a T7 promotor and a RBS into the pSB1C3 plasmide,which is the regulator.
This biobrick is in RFC 10 standards with: The prefixe (with ATG in frame): 5' GAATTC GCGGCCGC T TCTAGA TG GCCGGC and the suffixe (contain Stop in frame): ACCGGT TAAT ACTAGT A GCGGCCG CTGCAG 3'
AFP-lu1 constrcution in the pSB1C3 plasmid



