It was the first meeting in the CNRS of Marseille for the presentation of the iGEM competition by James and 2018 team members.
We attended a new presentation, more precise, of the competition and of its course. 2018 AMU team members developed their works by attribution, communication, funding, Human practices, modelisation and wetlab.
We began to suggest some project ideas. Propositions were numerous and dealt with various field. Recurring themes revolved about Health and Environment issues. We decided to further reading about these by groups.
15 subjects were pre-selected and we had to continue finding and working on bibliography. Besides, new members began to decide on what comities they wanted to take part (communication, finance, human practices, wetlab...).
We continued to do more researches to find the perfect project to the AMU iGEM team. Besides, we began to fill in funding files and to find out about public events in Marseille.
It was the D day! We had to choose our final subject of project. Among all of the ideas, two was selected to a final vote; the first one was developing a detection kit to diagnose Tuberculosis and the second one was finding a bioprocess to degrade lithium battery. With a margin of two votes, the final decision was the tuberculosis idea.
First of all, Dr Stephane Canaan, a specialist of Mycobacterium tuberculosis, made to us a presentation of the disease and gave to us some tracks to develop a detection kit. Then, we met "Semioz design graphique". He talked to us about some regulation for the logo in France, about design tools and presented to us the work ok Eiko Ojala, an Estonian illustrator and graphic designer. He thought that we should inspire us with this work to design our logo.
There was a first global idea about the logo: one healthy lung and the other one sicked. We could represent that in different colors. The human practices community realized the first survey about typical knowledge of people about tuberculosis. The financial community finished and sent the first financial files. Finally, we created our logo (we finished the meeting very late).
It was the day when we decided our name project. We were looking for a funny one. We voted for “I want to be free” in reference to the famous song “I want to be free” interpreted by the band Queen.
In order to find the technic to develop our detection kit, we read lot of articles to discuss about which one we could use.
In order to find the technic to develop our detection kit, we read lot of articles to discuss We received the famous iGEM kit. We did a skype with the Thessaly Greek team. During this call, we talked about Tuberculosis, about our different project and why our two teams have chosen to work on this disease. We sent to them our first survey to share around them.
We discussed our first meeting with the public which takes part the weekend before: the “Souk des sciences”. It allowed us to meet an old members of the iGEM team of Lyon but also to meet people working with tuberculosis-infected patients. We talked about the things we can improve as an explanation of the tuberculosis test for diagnosis and the survey for the public.
We announce on our social network the beginning of our crowdfunding campaign and we start the organization of our Meet-up.
We start to discuss about the dry Lab.
The collaboration with RAL Diagnostic is formalized and we are waiting for the equipment they sent to us. Moreover, we wrote the safety form and the project resum.
We discuss about the advance of our experimentations. We received the Pins with our logo.
The characterization of the chromoprotein is in progress, the construction of the TALE is done. We did a partnership with Wiloo.
We had some problem in the wet Lab (the sonication breaks the TALE, the clonage of the polymerase seems to be impossible). We ordered the tee-shirts of the team.
We made a return on the interview of Yannis and Jonnas for the Provence. We booked the flights to Boston and we worked on the design of the poster.
We discussed about the “Forum des Biotech” which take place in Polytech and where we did a presentation of the iGEM contest and of our project.
We set up the laboratory and installed the machines.
Methylases
Bibliographic research to try to find a solution to capture DNA specifically without damaging it.
Design of primers for PCR and PSR
Like many other isothermal amplification methods, the PSR method relies on the use of a DNA polymerase with strand displacement activity. Two primers with inverted sequences at their 5 'end are required to initiate the reaction. These inverted sequences were extracted from a botanical gene. The primers were developed with the aim of specifically targeting the Kat g and Rpob genes in Mycobacterium tuberculosis and Mycobacterium smegmatis.
The primers were designed based on the alignment of several published sequences of the Rpob and Katg gene (access numbers in GenBank are indicated in parentheses) in Mycobacterium smegmatis (CP001663.1) and Mycobacterium tuberculosis H37Rv (NC_000962.3 ) as well as different outgroup species and then the identification of two regions with a high degree of homology surrounding a more variable region using the ClustaIW alignment tool of the Bioedit software (BioEdit v7.0.5, Tom Hall Ibis Therapeutics ). The physicochemical characteristics of the different primers were established using the Primer3plus software (Primer3PlusvVersion: 2.4.2) while the specificity of the different primer pairs was evaluated in silico using the Primer-Blast software against the base of data nr / nt.
Test Strips
We performed all Biobricks we needed to carry out our experiment. We build three antifreeze, polymerase, TolA3, PIII-N1-M13, and specific methylase proteins, which will be used to detect the specific gen of M.tuberculosis and the human amylase.
TALs
We had a first meeting to establish the project's workflow. Bibliography: On how to design the RNA guide of Crispr Cas9 (clustered regularly interspaced short palindromic repeats-associated protein 9).
Methylases
Bibliography on methylase at the end of the week thanks to MIT hacking paris
PSR
The polymerases used for PSR must have a 5 '→ 3' DNA polymerase activity as well as a high strand displacement capacity, but must be devoid of 5 '→ 3' exonuclease function. For this we have firstly recover the sequences of Geobacillus stearothermophilus (CP008934.1) and Geobacillus thermodenitrificans (NC_009328.1) which have a high activity around 60° C and a strong strand displacement capacity. However, in order to eliminate their 5 '→ 3' exonuclease function, we aligned these sequences with the large E. coli klenow fragment which is a polymerase devoid of exonuclease activity, and then recover only the part that aligns.
We then added a prefix and a suffix to the sequence in order to be able to insert it in the Rfc vector 25 as well as a 6-his tag and a TEV sequence to be able to purify it.
Test Trips
We finished the bobriocks design and we moved in a new Lab and met all over research teams around us.
TALs
Second meeting Bibliography: On transcription activator-like (TAL) effectors
Our internal wiki is working, we begin to fill the notebook and all pages!
Lab
Cleaned-up the lab and unboxing all the material that Amidex financed.
Methylases
Comparison of the different methylases according to the DNA used during the PSR. To select the methylases, we composed via SnapGene the enzymes of human amyloid (chosen for PSR) and the mycobacterium sequences (smegmatis and tuberculosis). Then as the choice of enzymes was limited, I used the NEB Cutter V2.0 software. After a while, my choice is turned on HhaI-M for mycobacterium and NlaIII-M for amylase. We then optimized the sequences using IDT.
PSR
DNA extraction:
Total DNA was extracted using the NucleoSpin Microbial DNA Kit (ref: 740235.50) from a 10 ml of Mycobacterium Smegmatis culture. The extracted DNA was eluted with 100 μl of elution buffer and the DNA was stored at -20° C.
Polymerase Chain Reaction:
In order to test the specificity of our primers on Mycobacterium smegmatis, we performed a PCR.
The polymerase chain reactions (PCR) were performed in a SENSOQUEST automated thermal cycler in 25 μL of PCR mix containing 12.5 μl of Econo taq PLUS GREEN 2X Master mix and 0.1 μl of each primer (10 μM). The 16S rRNA gene was amplified using the pair of primers described in the previous part (IDT) and 1 μL of DNA template (11 ng / μl). An initial denaturation step of 5 min at 95 ° C was followed by 39 denaturation cycles of 1 min each at 95 ° C, a hybridization step of 30s at 55 ° C and a treatment of 60 s of elongation at 72 ° C. Holding at 72 ° C for 5 minutes allowed complete extension of the PCR products. Negative controls consisting of PCR mixing without DNA template were included in each run of PCR..
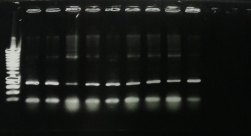
For the Katg gene, we obtained a band around 300 bp which corresponded well to the expected result (theoretical size 304 bp).
For the Rpob gene, we obtained two bands, one at 600 bp and another 800 bp. The expected size of the amplicon was 600 bp. So we have a nonspecific band. In addition we observed strong bands of small sizes corresponding to the primers. This means that our primers are too concentrated.
Polymerase Chain Reaction:
In order to increase the specificity of our primer pair for the Rpob gene, we performed a new dilution from the stock solution of our primers, then we carried out a gradient PCR with the following temperatures: 48.8 ° C, 52.8, 57.2, 65 ° C. The protocols used are the same as those used previously.
Results:
We obtained a single band at 600 bp for the following temperatures: 48.8 ° C, 52.8, 57.2. Our primers have therefore amplified specifically the Rpob gene.
No bands were observed at 65° C.
Production of Dna Polymerases
We did a ligation in a pSB1K3 plasmid at room temperature for 3 hours, and transformed the product into DH5α competent cells. We ran a colony-PCR. Verification on an agarose gel of the colony-PCR (ligation with our insert and psB1K3 plasmid). We launched starters on the confirmed positive colonies for the next day. We purified the DNA (starter) using a DNA purification miniprep kit, measured the DNA concentration on the NanoDrop and ran a digestion test and sent the product for sequencing.
Methylases
Command of the methylases.
Polymerase spiral Reaction
The PSR reactions were carried out in a 25 μl reaction mixture containing the following components: 1.0 μl Bst DNA polymerase large fragment (New England Biolabs), 2.5 μl 10 × ThermolPol reaction buffer (New England Biolabs, including 20 mM Tris-HCl, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Tween 20), 1 ul Betain 0.8 M,1 ul MgSO4 6 mM, 3,5 10 mM each deoxynucleoside triphosphate. The amount of primers needed for one reaction was 1 ulfor both Forward and Reverse for Rpob and Kat g. Finally, 1 ul of DNA template was added to the reaction tube. The PSR reaction was carried out for 60 minutes at 57°C, 61°C, 64°C, 68°C.
The results were visualized by electrophoresis as explained previously.
Results:
No band could be obtained. This negative result is explained by the fact that amplicons obtained by the PSR are much too big and can not be visualized on gel.
Lab
Preparation of SDS 10% and APS 10%: that will be used to make Western-blot and SDS-PAGE gels.
Polymerase spiral Reaction:
In order to evaluate the PSR under different conditions, we added 1ul of SYBR GREEN in our mix. The results were visualized using the CFX 96 Real Time Cycler thermocycler by measuring the fluorescence every minute.
We tested different DNA concentrations (10 ^ 5 to 10 ^ 2 copies / ul) and different temperatures (66° C, 63.8° C, 59.6° C and 58° C)
Results:
The best results were obtained for concentrations of 10 ^ 4 and 10 ^ 3 of DNA at 63.1 ° C and 59.6 ° C. However, the amplification was not optimal, because the fluorescence did not even increase by a factor of 2.
Polymerase spiral Reaction:
The PSR was carried out under the same conditions as above with the target Katg gene because it contains a restriction site of the enzyme KatG. Thus, after amplifications a band should in theory be observable by gel electrophoresis. We therefore digested the amplification products with Eco RI, however no band was observed.
Test Trips
CBD
Extraction of te CBD DNA into the Kit Plate. DNA purification and enzymatic digestion was performed into the plasmid. The plasmidaws used to do a PCR and a clean up, and the optic density was measured thanks to the Nannodrop , either a DNA concentration 254 ng/µl.
TALs Biobrick
Digestion of pSB1K3 with EcoRI and PstI, and then a DNA clean up with Monarch kit after digestion. We received the gene sequence from IDT containing the prefix and suffix. PCR-amplification and agarose gel electrophoresis of the two linear TALEs received from IDT.
Antifreeze protein
Polymerase chain reaction:
The PCR was used to amplify antifreeze Gblocks, we're using three proteins in order to determine which one will work better in different conditions. After the amplification a band appears on every sample by gel electrophoresis.
A digestion of these inserts and T7_RBS vector, was done using restriction enzymes.
Ligation and transformation: anti-freeze inserts were added to T7-RBS vector, using a ligase. after the ligation, the vector was introduced into E.coli 5DH∂ using a transformation. Cultures were incubated over night
TALs Biobrick
We ran a gradient-PCR to find the optimal primers hybridization temperature and verified it on an agarose gel.
Polymerase spiral Reaction:
So far all our results have been negative, we think this is due to the concentration of betain. So we started again the mother solution of betain to 10M. Moreover we have inserted in our manipulations a positive control resulting from an article. We used a 0.8M betain concentration, a DNA concentration of 10 3 copies and the reaction was carried out at 65 ° C for 70 min. In order to visualize a possible amplification, 1 μl of SYBR GREEN (1/20) was added.
Result
For the negative control (mix without DNA) no color change was observed. For the positive control, we observe a high intensity green color which means that the PSR has functioned. For our samples (rpoB) a slight color change was observed which means that the amplification was not optimal.
Test Strips
Blue Green Chromoprotein
A PCR on the Blue-Green Chromoprotein DNA fragment was realised to amplify the sample that we received by IDT. We verifyed the PCR product by Agarose gel elecrophoresis. The migration was performed during 25 minutes at 135 Volts, and was revelated with REDGel (BET), and the DNA concentration have been determined.
TALs Biobrick
Digested the gene with respective enzymes EcoRI and PstI, and then a DNA clean up with Monarch kit after digestion.
Design of primers for PCR and PSR
The PCR was used to amplify Polymerase Gblocks, we are using two proteins Geobacillus stearothermophilus (CP008934.1) and Geobacillus thermodenitrificans, we received the gene sequence from IDT. We ran a gradient-PCR to find the optimal primers hybridization temperature and verified it on an agarose gel. We have digested the gene with respective enzymes EcoRI and PstI, and then a DNA clean up with Monarch kit after digestion. We did a ligation in a pSB1K3 plasmid at room temperature for 3 hours, and transformed the product into DH5α competent cells. We ran a colony-PCR. Then we have made a verification on an agarose gel of the colony-PCR (ligation with our insert and psB1K3 plasmid). We had no band at the right size.
Methylases
Recovery of genes blocks by IDT, the sequences sent on twist took too long to arrive.
TALs Biobrick
We did a ligation in a pSB1K3 plasmid at room temperature for 3 hours, and transformed the product into DH5α competent cells.
Test strips
Amplification of the p3-N1, TolA3-CBD and Blue green protein DNA fragment was performed. An agarose gel electrophoresis have been made to verifiyed the insert size.
TALs Biobrick
We ran a colony-PCR on the positive (white)/negative (pink) colonies.
Test strips
An second PCR have been made in order to add a linker on each DNA fragment. That PCR was verified by agarose gel electrophoresis at 1%. We done a clean up on each DNA sample we obtained.
TALs Biobrick
-Verification on an agarose gel of the colony-PCR (ligation with our insert and psB1K3 plasmid). We launched starters on the confirmed positive colonies for the next day.
-Digestion of the plasmid containing the promoter T7 and RBS (BBa_K525998) with respective enzymes SpeI and PstI and both TALs but with XbaI and PstI enzymes, then we did a DNA clean up with Monarch kit after digestion. To finish we ran an overnight ligation in a psB1C3 plasmid at 16°C.
Test Trips
Fusion proteins constructions
A plasmid digestion was performed in order to insert each DNA fragment into a vector. The PCR products containing each target DNA fragment have been cleaned by DNA clean up Monarch Kit.
A SLIC methode have been performed to insert two DNA fragment into a vector in order to create a fusion protein.
5µl of each sample was used to carry out a transformation with DH5α.
Anti-freeze result
No bacteria survived. same digestion and ligation were preformed, but with a different mix
Specificity of the PSR Assay
SYBR Greenı can bind to double-stranded DNA, emitting green fluorescence under UV light. Experiments were also performed to determine the detection of rpoB PSR assay when SYBR Green was used to enable observation of positive reactions using a visual color change
The PSR assay specificity for M.smegmatis detection was evaluated by SYBR GREEN indicator colorimetric assay. the strains tested are : Vibrio cholerae, L..lactis, Lysteria monocytogenes, Burkholderia cenocepacia, Acinetobacter baumannii, E.coli 0157 H7, Salmonella typhimurium, Xenorhabdus nematophila, Caulobacter crescentus, Corynebacterium olae, Photorhabdus luminescens, Xanthomonas citri, Streptococcus mutans, M. smegmatis (C+) and Water (C-).
A color change was visualized for M.smegmatis, Lysteria monocytogenes and Burkholderia cenocepacia.
Sensitivity of PSR Assay
To compare the sensitivities of the PSR method and conventional PCR amplification, we conducted 10-fold serial dilutions of M.smegmatis genomic DNA (1.1ng/ul - 0,00011ng/ul) and subjected them to sensitivity testing. We tested primer for Rpob and Kat g.The detection limit for PCR was 0,11 ng/ul based on electrophoresis for rpoB and KatG gene.
TALs Biobrick
- We purified the DNA from a confirmed positive colony-PCR using a DNA purification miniprep kit and measured the DNA concentration on the NanoDrop and ran a digestion test.
- The ligation in psB1C3 was then transformed the product into DH5α competent cells.
Test Trips
Fusion proteins constructions
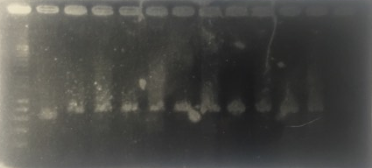
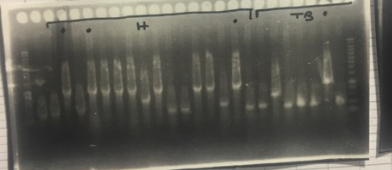
A screen PCR was realised to make sure that the TolA3-CBD and P3-N1-Green chromoprotein have inserted themselves into the plasmid. The results are shown in the following figure.(picture p24)
Recovery of strain of mycobacteria previously denatured at 90° C for 30 minutes: M. tuberculosis MC26230, M. avinum, M. BCG and M. marinum.
TALs Biobrick
We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies, the amplification didn’t work.
Test Trips
Fusion proteins constructions
A culture was performed on 3 colony of P3-N1-Green chromoprotein and 2 clones of TolA3-CBD.
The DNA sample from each selected clone were prepared by miniprep monarch kit and were sent to the sequencing.
Methylases
PCR to bind the methylase to the CBD.
Anti-freeze result
Transformation still not working. After reviewing old results, the amount of plasmid is changed.
TALs Biobrick
- We did an interne digestion on the purified colonies because the size of our insert is the same size of the mRFP of the plasmid psB1K3. Digestion on the purified sample with EcorI and NcoI from NEB, then was verified on an agarose gel and the results were negative.
- We recovered the transformation plates of the day 04.07 and ran another colony-PCR. We then launched starters from the confirmed positive colonies by an agarose gel.
Test trips
Fusion proteins constructions
The sequnecing results shown that the inserts TolA3-CBD and P3-N1-Green chromoprotein were absents into the plasmids.
A plasmid digestion (pSB1K3) was made with EcoRI and PstI restriction enzyme.
DNA clean up by monarch kit.
Anti-freeze result
After digestion and ligation, the culture is put into a transformation overnight
TALs Biobrick
We purified the DNA (starter) using a DNA purification miniprep kit, measured the DNA concentration on the NanoDrop and ran a digestion test and sent the product for sequencing.
Test trips
Fusion proteins constructions
A plasmid digestion (pSB1C3) was made with EcoRI and PstI restriction enzyme.
DNA clean up by monach kit.
Digestion of the PCR produts after the clean up with the T4 DNA polymerase: The T4 DNA polymerase was used to digerate the inserts (TolA3-CBD and P3-N1-Green chromoprotein) and the vector pSB1C3 during 2.30 minutes. The sample were introduced into ice for 10 minutes to stop the reaction. Transformation with DH5α strain cell.
Anti-freeze result
A screenPCR was used to verify the size of the plasmid to make sure that the plasmid is well ligated to the insert. And a culture was incubated overnight
TALs Biobrick
We recovered the transformation plates of the day 04.07 and ran another colony-PCR because it worked for only one TALE and verified it on an agarose gel.
Test trips
Fusion proteins constructions
A scren PCR was realised in order to verified the incorporation of the inserts into the plasmid.
The DNA sample from each selected clone were prepared by miniprep monarch kit and were sent to the sequencing.
Anti-freeze result
The culture that shown the must adequate size was chosen to do a sequencing, so a starter that contains antibiotics (chloramphenicol) was used to incubate overnight.
TALs Biobrick
We launched starters on the confirmed positive colonies for the next day.
TALs Biobrick
- We purified the DNA from a confirmed positive colony using a DNA purification miniprep kit and sent the product for sequencing.
- Digestion of the plasmid (psB1C3) containing the promoter T7 and RBS (BBa_K525998) and our TALE gene with respective enzymes SpeI and PstI and both chromoprotein chosen but with XbaI and PstI enzymes, then we did a DNA clean up with Monarch kit after digestion, measured the DNA concentration on the NanoDrop and ran a digestion test.
Methylases
Slic to link CBD to the different methylases.
Anti-freeze results
Mini-preps were performed to purify the plasmid. and sent to sequencing.
TALs Biobrick
We ran a ligation in pSB1C3 at room temperature, was then transformed the product into DH5α competent cells (didn't worked).
Test strips
Digestion of the plasmid (psB1C3) containing the promoter T7 and RBS (BBa_K525998) with PstI and PstI-HF a, then we did a DNA clean up with Monarch kit after digestion at 37°C. An hybridation have been made with the T4 DNA polymerase and a transformation with E.coli DH5α competent cells have been performed.
TALs Biobrick
Digestion of the plasmid (psB1C3) containing the promoter T7 and RBS (BBa_K525998) with SpeI and PstI-HF and our TALE gene with XbaI and PstI, then we did a DNA clean up with Monarch kit after digestion, measured the DNA concentration on the NanoDrop and did a ligation overnight at 16°C.
Test strips
We use the transformants DH5α to carry out a miniprep. First cwe inoculate these transformants into LB medium containing ampicilline at 37°C during 16 hours.
TALs Biobrick
We cloned the gene in pSB1C3 overnight and trasnformed it into DH5α competent cells.
Test strips
Miniprep
TALs Biobrick
- We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. We had non-conclusive results.
- We did digestion and ligation with a linear plasmid named psB1K3 with our gene overnight at 16°C.
- We did another PCR-amplification and agarose gel electrophoresis of the two linear gene received from IDT, to renew our DNA stocks.
TALs Biobrick
We cloned the gene in pSB1K3 overnight and trasnformed it into DH5α competent cells.
Test strips
We use the plasmids that are purified during the miniprep step to do PCR in order to add a linker on each gene fragments.
Clean up PCR.
SLIC : We used Cut plasmid (pSB1K and PSB1A3) to carry out a SLIC. A ration of 1 vector for two inserts have been calculated for each differents plasmids we used. An Hybridation have been made with the T4 DNA polymerase. The mix have been incubated at room temperature during 2.30 minutes and have been placed into ice for 10 minutes to stop the reaction. The transformation of 50µl of DH5α competents cells with 5µl of hybridation mix (insert/vector).
The culture have been incubate in a petri dish containing LB medium and Kanamycine or ampicilline antibiotics during a week-end on the a bench.
We ran a ligation in pSB1C3 at room temperature, was then transformed the product into DH5α competent cells.
Polymerase spiral reaction
Receipt of strains of denatured mycobacteria that have lost all pathogenicity. DNA extraction was performed from a 1ml pellet using the Nucleo Spin Microbial DNA kit. The DNA concentration was measured by Nano Drop . We performed a PCR to test our M. tuberculosis specific primers. and then we performed a PSR to test our M. tuberculosis specific primers. To analyze the PSR reaction product, the product was purified with a DNA purification kit, anddigested with PvuI restriction enzyme cutting sites found in the original sequence the digestion product has been visualized on agarose gel.
Methylases
Other week of SLICs to bind the methylase to the CBD.
Anti-freeze result
The result of sequencing was incorrect (empty vector). An other protocol was conducted (linear plasmid)
TALs Biobrick
We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. We had non-conclusive results.
Test strips
A lot of clones was observed on the petri dish.
A screen PCR with one Taq mix was performed to verified the presence of the inserts into the plasmids.
Storage at 4°C.
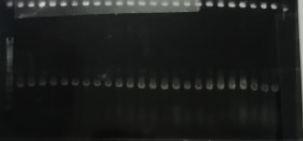
The results of the screen PCR are shown below.
Culture over night of the clones 1,4, 12 and 18 of the transdormants containing the pSB1A3 plasmid with the CBD-TolA3 insert.
TALs Biobrick
We digested the gene sequence with EcoRI and PstI for cloning in pSB1C3. We had the right band at the right size when migrated on an agarose gel then we cloned the digested gene sequence in pSB1C3 and transformed into DH5α competent cells.
Test strips
A SLIC have been made with the ratio 1 :2 :2 (vector/insert 1/insert 2), then we incubate the hybridation mix at room temperature during 2.30 minutes with the T4 DNA polymerase. The reaction have been stoped when we put the sample into ice for 10 minutes.
Transformation of 50µL of competents cells by heat choc with 3 to 5 µL of the inserts/vector mix.
TALs Biobrick
We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. Afterward, we prepared 3 starters on the confirmed positive colonies for plasmid purification.
Test strips
- Miniprep
- Verification of the presence of the insert into the plasmid by digestion with restriction enzymes .
- We digested the 3 clones containing TolA3-CBD ( clones 1,4 and 12) with two differents restriction enzymes such as EcoRI and PstI-HF. We carry out three differentes digestion for each sample. These digestions have been made during 1.30 hour at 37°C.
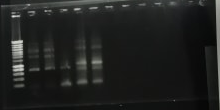
- The results are shown below
TALs Biobrick
- We did a digestion of verification and we didn't had the right band at the right size it had a size of a mRFP
- A gel extraction was done using a Macherey-Nagel kit, which allowed us to extract only the plasmid without the mRFP.
Test strips
- Digestion of pSB1A3-CBD-TolA3 with XbaI and pSB1-HF restriction enzyme. We digested CBD DNA fragment with XbaI and the TolA3 DNA fragment with PstI.
- Digestion of the plasmid pSB1C3 containing the T7RBS domain with SpeI and PstI-Hf restriction enzyme.
- DNA clean up by monarch kit
- Ligation : The T4 DNA ligase was used to ligate the insert TolA3-CBD and the T7RBS plasmid together. We put the T4 DNA ligase in contact with the insert/vector mix for 2 hours at room temperature.
- DNA clean up by monarch Kit
- Transformation of 50µL of competents cells by heat choc with 3 to 5 µL of the inserts/vector mix. Incubation at 37°C for 16 hours.
Test strips
- Miniprep
- Sequencing
- PCR of the blue-green chromoprotein and the blue chromoprotein have been made in order to add a linker on each DNA fragment. These linker have complementary base.
- Clean up PCR .
- Storage at -20°C
Polymerase Spiral reaction
To test the detection limit of the test for M. tuberculosis, we used genomic DNA from M. tuberculosis as a test object, and performed a series of 10-fold dilutions of the total DNA solution, generating in nucleic acids from 10^5 to 10^1 copies.
Methylases
An other week of SLICs.
TALs Biobrick
With the plasmid extracted we did a ligation for 2 hours at 25°C with our gene, that was then transformed into DH5α competent cells.
Test strips
Two differents SLIC have been made with the ratio 1 :2 :2 (vector pSB1A3 linear plasmid) ( /insert 1 (Blue chromoprotein )/insert 2 ( P3-N1)) and (vector psB1K3 linear plasmid)/ insert 1 ( Blue-green chromoproetin/ insert 2 ( P3-N1)), then we incubate these hybridation mix at room temperature during 2.30 minutes with the T4 DNA polymerase. These reaction have been stoped when we put the samples into ice for 10 minutes.
Transformation of 50µL of competents cells by heat choc with 3 to 5 µL of both inserts/vector mix.
Incubate 16 hours at 37°C.
TALs Biobrick
- We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies. We had no band at the right size.
- We used the SLIC/digestion method to clone our gene into a linear plasmid psB1K3, follow up with a transformation in DH5α competent cells.
Test strips
- Miniprep
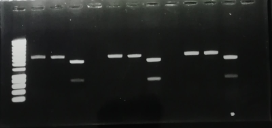
- A PCR colony have been made on each Blue Green –P3-N1 and Blue-P3-N1 clones.The results are shown below.
- Incubation af each clones at 37°C for 16 hours.
TALs Biobrick
- We did an other PCR-amplification and agarose gel electrophoresis of the two linear gene received from IDT, to renew our DNA stocks and we did a digestion with XbaI and PstI to clone into a plasmid containing T7 RBS that was cut with SpeI and PstI enzymes.
- We recovered the transformation plates of the SLIC and ran a colony-PCR on the supposedly positive colonies, 1 of 2 genes had the correct band size so we launched a starter.
Test strips
- Clean up PCR of the Blue-Green chromoprotein-P3-N1 and the Blue chromoprotein-P3-N1 have been made.
- Sequencing
TALs Biobrick
- We sent the miniprep product for sequencing.
- We did a ligation of the digested gene sequence in pSB1C3 with T7 RBS and transformed into DH5α competent cells. (Julie)
Test strips
Sequencing results : We have no insert between the prefix
TALs Biobrick
We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies.
Methylases
Transformation, ligation and digestion of the methylases.
TALs Biobrick
- We recovered the sequencing results that were positive (SLIC).
- We launched 6 starters of colonies that had the correct band size after PCR-Screen.
TALs Biobrick
- We did a digestion of verification on the minipreps sample, then sent the minipreps product for sequencing.
- We did multiple transformation to renew our stock with the RED, Bleu-Green, Bleu chromoprotein and the the plasmid (psB1C3 T7 RBS) containing both of our genes of interest.
TALs Biobrick
- We recovered the sequencing results that were positive (our gene + psB1C3 with T7 RBS)
- Digestion of the plasmid containing the promoter T7 and RBS (BBa_K525998) and our gene (TAL) with respective enzymes SpeI and PstI and both chromoproteins (Red (BBa_K592012) and Bleu (BBa_K1033928)) but with XbaI and PstI enzymes, then we did a DNA clean up with Monarch kit after digestion. To finish we did a ligation for 2 hours at 25°C and to finish they were then transformed into DH5α competent cells.
TALs Biobrick
- We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies.
- We launched 5 starters of colonies that had the correct band size after PCR-Screen.
Specificity PSR test
Realization of PSR with different DNA concentrations of M. tuberculosis for a test of sensitivity The specificity of the PSR Assay for M.tuberculosis Primers was tested.
Methylases
Slics finally worked, but we lack of time.
TALs Biobrick
- We sent the minipreps product for sequencing and launched a transformation in BL21 for the production.
- Digestion of the plasmid containing the promoter T7 and RBS (BBa_K1033928) and our gene (TAL) with respective enzymes SpeI and PstI and cjBlue green chromoprotein (BBa_K592011) but with XbaI and PstI enzymes, then we did a DNA clean up with Monarch kit after digestion. To finish we did a ligation for 2 hours at 25°C and to finish they were then transformed into BL21 competent cells.
- AMY1 DNA was obtained from sputum. DNA extraction was performed from a pellet of 1ml of saliva using the Nucleo Spin Microbial DNA kit. We performed a PCR to test our AMY1 specific primers and then a PSR to test our AMY1 specific primers.
TALs Biobrick
- We launched 3 starters of 10mL for the productions test.
- We recovered the transformation plates and ran a colony-PCR on the supposedly positive colonies and launched 1 starter of a colony that had the correct band size.
TALs Biobrick
- We recovered the sequencing results that were positive, psB1C3 containing T7 RBS+ TAL (our gene)+ Chromoprotein
- We sent the minipreps product for sequencing
- We induced till an OD of 0,5/0,6 at 37°C with a rotation of 850rpm then we added IPTG and incubateur during 3h at 30°C. 1mL of the cell culture was used to check the production on SDS page, by incubating the cell culture for 5 min at 95°C.
TALs Biobrick
We launched 3 starters of 10mL for the productions.