The design of Xylencer was a lively process which started in February 2019 and did not stop until now. The project went through countless iterations, motivated by discussions we had with supervisors, stakeholders, experts and within the team. On this page, we proudly present the status quo of Xylencer. Our project consists of four pillars: Detection, Delivery, Remediation, and Spread. Each of the single pillars can stand and represent a project on its own. However, we believe the combination of all four pillars is most effective to fight off Xylella fastidiosa. Read further to get to know more about the principles and ideas behind the design of Xylencer, which changes the project has gone through, and how we scientifically addressed the challenges ahead of us.
Detection
The first pillar of Xylencer is detection of X. fastidiosa. Since its first detection in Europe in the south of Italy in 2013, the pathogen has been detected throughout the Mediterranean, reaching from Portugal until Israel [1; 2]. A major bottleneck is the surveillance of X. fastidiosa occurrence via reliable and easy-to-use detection methods (see entry on Martijn Schenk in Human Practices). Current detection methods are quite labor-intensive and costly and therefore don’t allow large scale monitoring of the pathogen’s spread. Xylencer tackles this problem by developing an automated tool for in-field testing of X. fastidiosa in insect vectors using Loop-mediated isothermal amplification (LAMP). Insects are attracted by a plant mimic on which they feed and thereby transfer X. fastidiosa cells. In the plant mimic the LAMP reaction takes place detecting the pathogen in an automated manner. This way, Xylencer will help to better monitor the pathogen’s spread throughout Europe, enabling the more efficient coordination of control measures to combat the disease.


Design Considerations
-
Why use LAMP for detection? arrow_downward
LAMP is a method similar to PCR and allows specific amplification of a target sequence using six primers. The advantage over standard PCR is that the amplification takes place at a constant temperature between 60 – 65 °C. The isothermal reaction conditions, combined with the specificity and sensitivity, allow LAMP to be used as an automated tool for pathogen detection [3]. As iGEM is a competition in the field of synthetic biology, we first considered a SynBio approach to develop a detection tool. Especially since we already work on a receptor detecting X. fastidiosa quorum sensing molecules and transducing a signal (see the DSF sensing circuit page). However, in the end we decided on using LAMP for the following reasons. First, LAMP is already an established method for detecting various diseases in humans and plants [4; 5]. Second, in-field applications of LAMP are on the rise and deliver results as good as, or better than, PCR [6]. Third, LAMP avoids biosafety issues concerning the use of GMOs outside laboratory conditions. Indeed, the use of GMOs, especially in outside-lab applications, should always undergo a cost-benefit analysis: Does the risk associated with using synthetic biology and GMOs for the detection of X. fastidiosa counterbalance the benefits of this particular approach? Are there feasible alternative solutions for the same problem associated with lower risk? In our opinion, LAMP does portray such an alternative solution over the use of synthetic biology.
-
What does the ideal tool look like? arrow_downward
During our Human Practices, it became clear that surveillance of the pathogen’s spread is challenging, as infected plants stay asymptomatic over a long period of time, or never show symptoms. Therefore, the monitoring of X. fastidiosa movement by manually sampling plants in yet seemingly unaffected areas throughout the Mediterranean is impossible to conduct. Testing insects instead seems more feasible and effective, as wherever insects carry the pathogen, the disease will show up sooner or later. With Xylencer we designed a detection tool to test the insect vector carrying the pathogen instead of sampling plants themselves.
Within the Xylencer project we designed a plant mimic attracting the insect vectors that they can probe and feed on. This way, potential X. fastidiosa cells attached to the insect’s feeding apparatus, the stylet, would remain in the feeding solution. LAMP reagents are added in an automated manner to the feeding solution resulting in a readable colour output.
In order to perform insect experiments and develop an automated insect-sampling detection tool, we looked into how to attract and catch insects. Actively catching insects with nets does not allow large scale monitoring of pathogen spread considering the hours of labour involved. Therefore we considered the possibility of attracting and trapping the disease vectors. However, for the most prominent X. fastidiosa vector in the US, the glassy-winged sharpshooter, no pheromones for attraction are known . Also, sticky traps were already tested and found to be inefficient [7]. An alternative to attract insects is the usage of specific light wavelengths. However, these are not yet known for neither the spittlebug (the most prominent vector in Europe), nor the glassy-winged sharpshooter. Even though facilities to conduct light attraction experiments were available, we were not able to catch enough insects to perform light attraction assays.
Insects used in the experiments conducted in Xylencer were caught in the wild as in-laboratory breeding of the spittlebugs lasts over a 100 days [8].
-
How to test our tool? arrow_downward
Previously designed LAMP primers are used to assess the sensitivity of the detection tool [5]. Regarding the plant mimic, different containers have been evaluated for positive feeding behavior of the insects. Additionally, insect mediated transmission of E. coli expressing adhesion proteins on their cell surface has been assessed.
For more information visit the detection device results page.
Delivery
Phage therapy is a very promising technique for combatting bacterial infections, but application in agriculture suffers from problems in delivery of phage particles. In the second pillar of Xylencer we create a Phage Delivery Bacterium (PDB) to overcome these issues.
In our meeting with Britt Koskella, evolutionary biologist at UC Berkeley, USA, we got aware of the difficulties phage therapy is facing in agricultural applications. The initial phage titer and the location of their bacterial host are currently limiting the efficiency of phage therapy in crop plants. Also, environmental factors, such as pH, temperature and most importantly UV light, drastically impact the effectiveness of phages to control bacterial disease.

With the introduction of the Phage Delivery Bacterium (PDB) we overcome these issues. The PDB carries the Xylencer phage genome as a plasmid. This way, the phages are protected against the impact of high temperatures and UV light. Instead of spraying the phages on crops or inoculating infected plants with the pure phage particles, the PDB is injected into the plant’s xylem tissue. The proliferation of the Xylencer phage is linked to a mechanism sensing the presence of X. fastidiosa. If the pathogen is present, the repression is released and the Xylencer phage proliferates in the PDB until cell lysis. Released phages are now present in high titers and in direct proximity of X. fastidiosa.
Go To Resultsnavigate_next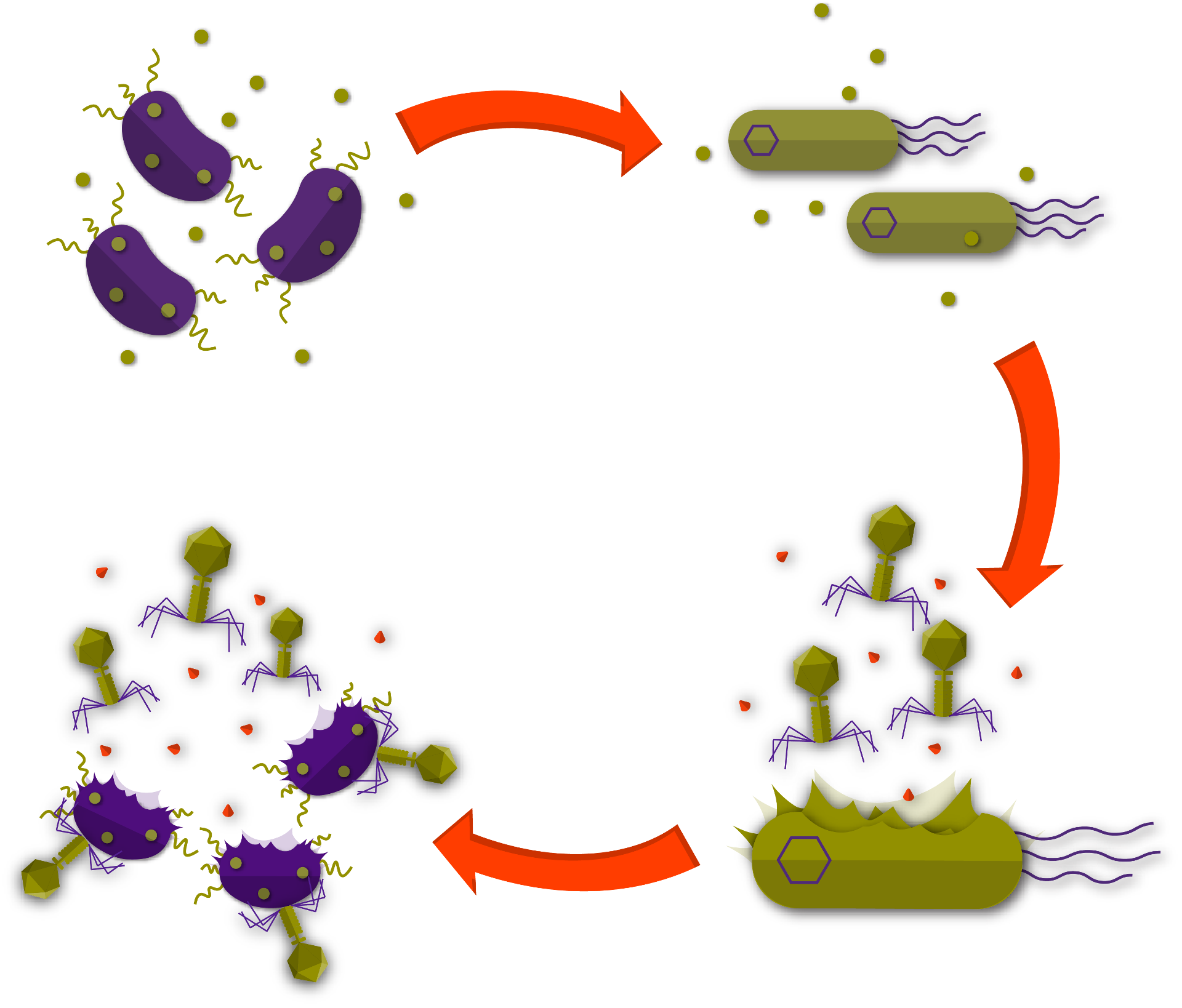
Design Considerations
-
Practical application/injection of our therapy arrow_downward
Overall, we considered two main methods of applying our PDB. Option one was equipping insects with the PDB, which in this case, would contain chitin binding proteins. Option two was injecting the PDB into the tree. Due to difficulties with both rearing and attracting the insect vectors, we eventually chose to inject the PDB.
The PDB will be injected into the xylem using an already existing apparatus specifically designed for this purpose [41]. The method does not require the farmer to have any special equipment and can therefore be applied easily in many locations. Besides this advantage, it is also inexpensive and does not cause harm to the plants. Reapplication is therefore also relatively easy.
This apparatus consists of an injector element, which can be inserted in a hole drilled in the trunk, combined with a latex tube. This latex tube can be filled with a solution containing the PDB. Due to negative pressure this solution will be sucked up into the xylem [41]. We felt like this method of applying would make our therapy very feasible for farmers.
-
Xanthomonas as PDB for the Xylencer phage arrow_downward
To serve as the PDB for our Xylencer phage, the chassis organism must fulfill specific requirements. First, the organism of choice must possess a cell metabolism sufficient to serve as a platform for X. fastidiosa phages to proliferate. Second, the PDB has to maintain a healthy state within the nutrient poor plant xylem to be able to produce high titers of phages. Third, chassis organisms may not have virulent properties on their own to reduce the risk associated in using them. We chose Xanthomonas as our PDB as it fulfills most requirements for the Xylencer PDB. The Xylencer phages have been tested successfully on various subspecies and strains of its host pathogen, as well as on few Xanthomonas species found in the phage’s natural proximity [9; 10]. We concluded that the X. fastidiosa phages might not be able to infect a wide range of Xanthomonas species, but that their cell metabolism is sufficient for the phages to proliferate. To the best of our knowledge these phages have not been tested on other, more prominent, chassis bacteria. Further, Xanthomonas is known for its ability to colonize the xylem tissue of plants [11; 12]. Maintaining a healthy and productive state should therefore not pose a problem. However, Xanthomonas is known as a largely pathogenic genus, with a few reported non-pathogenic strains. The literature behind these non-pathogenic strains is not extensive enough for us to label the reported strains as non-pathogens.
To increase confidence in the non-pathogenic status of these strains, we performed an extensive bioinformatic analysis. 1372 Genomes were retrieved from the NCBI database and reannotated with protein domains. Literature research was performed to curate a high-quality dataset of 104 genomes with experimentally verified pathogenicity. This dataset was used to train a random forest machine learning model. With the information from this model, we selected a set of non-pathogenic strains that could reliably be separated from the pathogenic strains, increasing confidence in their non-pathogenic status. This lead to the selection of Xanthomonas arboricola CITA 44 as our PDB.
For more information visit the pathogenicity analysis results page and the entry on horizontal gene transfer on the Biosafety page.
-
Sensing Xylella fastidiosa's presence arrow_downward
Even though Xylencer relies on having high phage titers to be more effective against X. fastidiosa, we do not aim to unnecessarily spread phages in the absence of the pathogen. By use of a genetic circuit, the proliferation of the Xylencer phages is dependent on sensing of X. fastidiosa presence. This way the spread of genetically engineered phages in the environment is limited to situations when our PDB encounters the pathogen. Also, the circuit enables the PDB to produce high titers at the pathogen’s actual location within the plant. DSF comprises the quorum sensing molecules used by the pathogen to regulate its behavior and are spread throughout the plant’s xylem if infected by X. fastidiosa [13]. A DSF receptor on the outer membrane of the PDB is linked to a riboswitch regulating the ON-/OFF-state of the genetic circuit. Upon X. fastidiosa presence, the riboswitch is switched to the ON state, leading to an activation of the downstream gene circuit and subsequent phage proliferation.
To test the system, GFP was placed under control of the riboswitch. This system was tested under induction of the sensing mechanism with multiple DSF variants in different concentrations. In parallel, we tested the functionality of the riboswitch independently of the DSF sensor. Ultimately, the GFP would be replaced to link the X. fastidiosa sensor to the complete circuit.
For more information visit the sensing X. fastidiosa results page.
-
Tight control of phage production arrow_downward
To prevent the unintentional release of Xylencer phages and to prevent lysis of the PDB before encountering X. fastidiosa, the proliferation of the phages needs to be tightly controlled. We achieve this by using CRISPR interference to target crucial phage operons and in this way repress the phage’s proliferation. To release the phage repression upon X. fastidiosa presence, an Anti-CRISPR (Acr) has been coupled to the beforementioned sensing mechanism. This way an ON-/OFF-switch for the proliferation of the Xylencer phages has been generated, linked to X. fastidiosa sensing mechanism. We adjusted the expression levels of the Acr allowing us to fine-tune the system and obtain ideal levels of phage proliferation repression and re-initiation.
To test and fine-tune the gene circuit we performed plate reader assays measuring GFP. CRISPR interference represses GFP expression, while Acr induction restores the GFP signal. Further, the system was tested in a multiplex manner on phage lambda promoters controlling GFP and RFP expression. Next we used a phage lambda plaque assay to test whether phage proliferation could be controlled.
For more information visit the phage repression results page.
-
Timed Kill-Switch arrow_downward
The Xylencer PDB will not be contained in an enclosed environment but ought to be applied in orchards and fields. That’s why we designed an induced lethality mechanism, mostly referred to as kill-switch. Upon X. fastidiosa encounter, the PDB is lysed from within by the proliferating Xylencer phage. However, in a situation in which the pathogen is not being perceived by the sensing mechanism, a kill-switch sets in to stop the PDB from proliferating. This kill-switch relies on a toxin-antitoxin system which is linked to the oscillating mechanism of Synechococcus elongatus [14], slowly increasing the concentration of toxins in the cell. This way, if the PDB has not encountered X. fastidiosa and produced Xylencer phages, lethality is induced.
To test the design of this genetic circuit, the oscillator mechanism of S. elongatus will be expressed in E. coli. After induction, fluorescence intensity should increase with an oscillatory tendency. To do so, 96 wells-plate experiments were conducted, where the OD and the fluorescence were monitored for 72 hours. After that, and after visualizing a successful increase in fluorescence, the well-studied mazE/mazF toxin-antitoxin system will be introduced. When bacterial death is induced, a decrease in OD will be seen. In this manner, we could test the functionality of the kill-switch linked to the oscillating system.
For more information visit the kill switch results page.
Remediation
After delivery of the phages, the next step in Xylencer is to cure the plants. In the third pillar, remediation, we aim to clear out all X. fastidiosa from the plant. Although phage therapy is extremely promising, there are some difficulties associated with it. To be effective, phages must be present at the right concentration at the location of the bacterial infection. This is difficult for X. fastidiosa, as it spreads aggressively throughout the entire xylem system of plants [15]. Additionally, low population density, low replication rate or resistance acquisition of the bacteria reduce phage effectiveness [16]. As a result, it is often not possible to clear out all pathogenic bacteria by bacteriophage therapy. These drawbacks were confirmed during our conversation with prof. Britt Koskella, expert in interaction of phages with the plant’s microbiome. Xylencer therefore applies a second strategy to combat X. fastidiosa. We form an alliance with the plant, by artificially triggering a systemic plant immune response and as a result, clearing the pathogen out of the plant’s system more effectively.



Bottom: Phage mediated lysis of X. fastidiosa releases both new phages and MAMPs that go on to trigger the plants immune system.
Design Considerations
-
Lytic or lysogenic phages? arrow_downward
Bacteriophages can infect their bacterial hosts using two different life cycles: lytic or lysogenic. During the lytic cycle, phages proliferate within their host ultimately lysing the bacterial cell. In the lysogenic phase, phages integrate their DNA into their hosts genome where they stay dormant. Upon change of environmental or the host’s conditions, the dormant phages excise their genome and switch to the lytic phase [17]. Temperate phages can follow both life-cycles, while strictly lytic, or virulent, phages only follow the lytic cycle [18].
The use of lysogenic phages in phage therapy entails various risks and uncertainties. First, during the lysogenic phase, phages interact with the bacterial host in a process called lysogenic conversion. This can lead to an altered or even enhanced pathogenic phenotype of the bacterial host [19]. Second, lysogeny confers the host resistance against infection of other phages, therefore reducing phage therapy efficiency. Third, general transduction, the transfer of bacterial DNA in between various hosts, is mostly mediated by temperate phages as compared to strictly lytic phages [18]. An increasing rate of bacterial DNA exchange favors the development of traits such as phage resistance and can enhance virulence. In contrast, strictly lytic phages are more effective as they do not have a dormant state and proliferate and lyse their host. Besides these benefits of the lytic cycle, the use of virulent phages is also safer as lysogenic conversion and general transduction are not present or less likely. For X. fastidiosa, both lytic and lysogenic phages have been characterized [9, 20]. Due to safety considerations, we chose to use strictly lytic phages for the Xylencer phage therapy.
-
How do we avoid phage resistance? arrow_downward
Bacteria are highly flexible organisms, able to adapt to various outer influences and threats. In the medical sector, the appearance of multi drug resistant bacteria poses a problem as pathogens develop resistances against commonly used antibiotics [21]. In a similar way, bacterial pathogens can form resistance against phage therapy, for example by losing structures on their cell surface.
To avoid X. fastidiosa forming resistance against the Xylencer phages, we apply two different strategies. First, phages will be applied in mixtures of at least two phages which is referred to as a phage cocktail [22]. Differing mechanisms of infection make it less likely for the target bacteria to evade phage therapy. Further, phage cocktails ensure a broader activity range under varying conditions and could impact more bacterial types. If the immunity type between these phages differ, they could also infect bacteria containing prophages in their genome [18]. Second, these phage cocktails will be applied in an alternating manner. After application of a phage cocktail over a certain period of time bacterial resistance might occur anyway. Exchanging the phage cocktail in place for phages with different infection mechanisms might restore the pathogen’s susceptibility. Currently, there are four strictly lytic phages described for X. fastidiosa, meaning that we could apply two different cocktails consisting each of two phages [9]. While four phages are sufficient to apply alternating phage cocktails, the need for more phages is obvious. Also, more research on the phage infection mechanisms needs to be performed [10]. So far, X. fastidiosa pili were found to be necessary for phage infection but not sufficient [9; 10]. The phage cocktails in combination with the artificial activation of the immune response represent promising approaches to lower the development of bacterial resistance against the Xylencer phages.
-
How does X. fastidiosa evade the plant immune response? arrow_downward
The plant’s innate immune system relies on the recognition of an intruding organism and a subsequently triggered defense response. Plants recognize intruders through receptors binding conserved pathogen associated molecular patterns (PAMPs), also referred to as MAMPS, such as flagellin, lipopolysaccharides (LPS), chitin and many more [23]. X. fastidiosa manages to thrive in the xylem as it camouflages the part of its LPS that is recognized by the receptors with a long O-antigen chain. Thereby the pathogen avoids recognition and subsequent PAMP triggered immune (PTI) responses of the plant. To see the effect of PTI if triggered, purified LPS, where the immune activating part is exposed, or LPS without the long O-antigen chain were inoculated into the xylem. Both were shown to artificially trigger systemic defense responses strong enough to prevent infection by X. fastidiosa in plants [24].
-
How to activate the plant immune response against X. fastidiosa? arrow_downward
In Xylencer, we aim to equip the phages with a gene encoding for flg22 which entails 22 conserved amino acids at the N-terminus flagellin of Pseudomonas syringae [25]. Flg22 is a well described PAMP and was shown to activate the same defense signaling pathways as LPS which were effective against X. fastidiosa [26]. Furthermore, it is known to act as a plant immune response elicitor in a wide range of plants affected by X. fastidiosa. Also, flg22 is a peptide, as compared to other PAMPs such as LPS or chitin. Introducing a gene encoding for flg22 is considerably easier than introducing the enzymes active in the pathway of chitin and LPS synthesis. Upon phage infection, the pathogen’s cell metabolism will be highjacked in order to express phage genes, including the flg22 introduced by the Xylencer phages. After lysis of X. fastidiosa, flg22 will be released into the xylem. By artificially triggering a systemic plant immune response we will complement phage therapy in clearing out X. fastidiosa of infected plants.
-
Why activate the plant immune response? arrow_downward
During the design process, multiple second methods to combat X. fastidiosa, besides the lytic cycle of phages, were considered such as the release of toxins and biofilm destroying enzymes.
There are toxins that could target X. fastidiosa, such as conventional antibiotics or antibacterial peptides. One problem with these compounds is that they are often not specific and would also target the microbiome of the plant. This microbiome is important for plant health, so it is best to disturb it as little as possible [27]. Although the exact mechanism is not understood, activating PTI has no, or very little, detrimental effects for the microbiota [28]. Besides the lack of specificity, resistance could arise against these compounds. A preventive measure for resistance development is to avoid exposure to low concentrations of the active compound [29]. Within our application, the concentrations of exposure can vary substantially, facilitating the occurrence of resistance. Furthermore, genes encoding antimicrobial compounds could be transferred to other bacteria by general transduction, having unforeseeable effects on the microbiome. This is not the case for PAMP encoding genes as they do not confer any selective advantage. Due to these reasons we decided to activate the natural immune responses of the plant instead of including antibacterial compounds in our design.
X. fastidiosa forms biofilms both in the plant and on the insect vector as part of its pathogenic life-cycle [15; 30]. Enzymes to cleave EPS, a principle component of biofilms, are known but have not been tested yet on X. fastidiosda biofilm [31]. A previous iGEM team, Aix-Marseille 2017 worked on biofilm-degrading enzymes to tackle X. fastidiosa. We chose not to pursue this route due to the following two reasons. First, it is difficult to study the biofilm of X. fastidiosa as it grows extremely slow under laboratory conditions [32]. Second, as biofilms are species specific, it is not possible to work with model organisms and transfer the knowledge. The compounds in the biofilm of X. fastidiosa are not completely characterized [33]. Our early approach to predict biofilm degradation using bioinformatic analyses was therefore not feasible either. However, if biofilm degrading enzymes were shown to work on X. fastidiosa biofilm, these enzymes could potentially be a valuable addition to Xylencer.
-
How to test our MAMPs? arrow_downward
The effect of flg22 xylem inoculation was tested on a pair of model organisms in plant pathology. The pathogen Xanthomonas campestris pv. Campestris (Xcc) has a similar pathogenic life-cycle compared to X. fastidiosa. However, it is not considered a quarantine organism itself which makes it easier and safer to work with. Cabbage (Brassica oleracea) is a suitable host plant for X. campestris infections, as it shows clear symptoms and xylem inoculation is relatively easy due to the leaf structure and size. Plants were infected with Xcc until lesions appeared and subsequently xylem inoculated with flg22. To quantify the effects of flg22 treatment, the reduction in lesion size was measured.
For more information visit the plant immune response results page.
Spread
X. fastidiosa can infect over 350 crops and wild plant species, many of which do not show symptoms over a long period of time, if not for ever. This way, the pathogen has a tremendous hidden reservoir from which infections can spread continuously. To eradicate the disease for good, we equip the Xylencer phages with the ability to spread, making use of the same spreading mechanism that X. fastidiosa applies. This will allow the phages to reach X. fastidiosa reservoirs in the near proximity of farms and prevents reinfection of farm crops.

Design Considerations
-
How to effectively administer phage therapy? arrow_downward
X. fastidiosa can spread easily to different plants and fields via its insect vector and affects large areas of both cultivated and uncultivated land. Applying phage therapy to every plant individually would be laborious, if not impossible. Additionally, asymptomatic plants in the field’s surroundings would evade treatment and act as reservoirs for reinfection. First, based on the knowledge that insects also transfer plant viruses, we looked into natural mechanism of insect-virus interactions [8]. These mechanisms turned out to be very specific mutual interactions between insects feeding on particular plant species and associated viruses. However, it turned out no such interactions were known to literature for the most prominent X. fastidiosa vectors, so we searched for alternatives. We found the solution to the lack of potential interactions for the vectors in the very organism we are working to eradicate. X. fastidiosa has the ability to spread through insects by using adhesion proteins to bind to the feeding apparatus of insects, named stylet. These adhesion proteins primarily interact with chitin which is an abundant polysaccharide in insect exoskeletons, including the stylet [34]. In Xylencer, we fused these adhesion proteins to the capsid proteins of the phages. This way, the Xylencer phages are able to spread from plant to plant by the same insect vectors that spread X. fastidiosa. Treatment of a few plants with the phage delivery bacterium (PDB) could cure an entire plantation from disease. Further, asymptomatic and wild plants would not evade treatment any longer. Ideally, the phages stay in an area as long as the pathogen persists due to constant re-infection of newly emerging X. fastidiosa infestations. As soon as the pathogen is eradicated the phages lack a host and start degrading after 10 – 12 weeks [10]. This way, Xylencer provides a long-term solution for infested fields and areas by copying the pathogen’s own spreading mechanism.
-
Choice of model phage arrow_downward
To test the strategy described above, we needed a model phage similar to the X. fastidiosa's phages. Sequence analysis of the different phage genomes showed the highest similarity between the capsid morphology of the E. coli phage lambda and the X. fastidiosa phages Sano and Salvo [9]. Phage lambda's capsid consists of homo multimers of capsid proteins that are stabilized by a net of decorator proteins. Display of proteins on phage lambda, used often in phage display, is achieved by fusing the protein of choice to the decorator protein [35]. C-terminal fusions were shown to be effective at accommodating relatively large proteins [36]. We chose phage lambda as our model phage for two main reasons: the relatively high similarity between phage lambda and the X. fastidiosa phages, and the existence of proven protocols for high-capacity fusions.
-
Choice of adhesion proteins arrow_downward
Successful spread of our Xylencer phages does not solely rely on the ability to bind to the insect vectors’ stylet. Additionally, the release of the binding is crucial to be transmitted to more plants in the surrounding. Therefore, the goal was to create a library of proteins with different adhesion ability to test for optimal spreading abilities of the Xylencer phages. The first chitin binding proteins we looked at where the ones used by X. fastidiosa itself. During our Human Practices we got aware of the dual functions of these pathogen derived proteins in virulence and disease formation (see Rodrigo Almeida's entry at Human Practices). In response, we expanded our adhesion protein library. We added chitin binding proteins described in literature which do not play a role in pathogenicity and have been used for protein fusion before. In this way, we generated an adhesion protein library of potential capsid fusion candidates for future testing of phage transmission efficiency.
To test for the adhesion ability, we performed adhesion assays using chitin and chitin coated beats. Adhesion proteins of our library were recombinantly expressed and purified. After inoculation with chitin, the proteins were washed off and their abundance measured and compared to a non-chitin binding control protein.
For more information read the results page of the adhesion library.
-
Phage Engineering Approach arrow_downward
To engineer phages, various methods can be applied. Most of these methods rely on using the phage’s natural host as a phage engineering and proliferation platform [37]. This poses a problem in the case of X. fastidiosa phages. The pathogen is extremely slow growing under laboratory conditions resulting in engineering and proliferation of the phages to be very time consuming and low yielding [32]. Additionally, X. fastidiosa is listed as a quarantine organism in Europe making it more difficult to obtain the pathogen and work with it as sophisticated laboratories are needed. Also, the chance of unintentional X. fastidiosa spread is higher. As a result, we focused on exploring phage engineering and proliferation methods independent of the pathogenic host.
For this, we apply different methods for the engineering and proliferation of the X. fastidiosa phages. In yeast-based assembly, the phage genome is split into various fragments, including a fragment containing the capsid-adhesion protein fusions [38]. These fragments are transformed into Saccharomyces cerevisiae where the endogenous homologous recombination mechanism assembles the fragments. The obtained engineered phage genome can then be transformed into E. coli which leads to the first production of phage particles [37]. Alternatively, the engineered phage genome can be used in cell-free transcription-translation (TXTL) mixtures to obtain assembled phages [39]. This way, the Xylencer phages can be engineered and proliferated without the use of X. fastidiosa itself.
For more information see the phage engineering results page.
-
Spatial Spread Model arrow_downward
The success of Xylencer depends on the range of spreading of the phages. Besides the adhesion ability of the fused chitin binding proteins, the spread further depends on ecological factors. The prevalence of X. fastidiosa itself is important as the host will serve as the on-field proliferation platform of the phages. Also, the abundance of insect vectors, their feeding behavior, and travel range affect the efficiency of Xylencer phages to eradicate X. fastidiosa. As we cannot collect data on these factors in the field, we designed a spatial spread model.
To make this theoretical model, we first modeled the phage-bacteria interaction of X. fastidiosa and the phages. Next, we looked at existing models describing the spatial spread of X. fastidiosa and recreated these to gain insights in how the pathogen spreads itself [40]. At last, we were able to combine the phage-bacteria interaction and the X. fastidiosa spatial spread models. The result is a theoretical model for the spread of the Xylencer phages and their effectiveness at combatting X. fastidiosa. This model helps us to estimate the effectiveness of our phage therapy and the range of phage spread. This data is crucial to plan the future application of Xylencer phages in the field.
For more information see the spatial spread results page.
-
References arrow_downward
- Saponari, M., Boscia, D., Nigro, F., & Martelli, G. P. (2013). Identification of DNA sequences related to Xylella fastidiosa in oleander, almond and olive trees exhibiting leaf scorch symptoms in Apulia (Southern Italy). Journal of Plant Pathology, 95(3.
- Sicard, A., Zeilinger, A. R., Vanhove, M., Schartel, T. E., Beal, D. J., Daugherty, M. P., & Almeida, R. P. (2018). Xylella fastidiosa: Insights into an emerging plant pathogen. Annual review of phytopathology, 56, 181-202.
- Zhang, S. Y., Dai, D. J., Wang, H. D., & Zhang, C. Q. (2019). One-step loop-mediated isothermal amplification (LAMP) for the rapid and sensitive detection of Fusarium fujikuroi in bakanae disease through NRPS31, an important gene in the gibberellic acid bio-synthesis. Scientific reports, 9(1), 3726.
- Poon, L. L. M. et al.(2006) ‘Sensitive and Inexpensive Molecular Test for Falciparum Malaria: Detecting Plasmodium falciparum DNA Directly from Heat-Treated Blood by Loop-Mediated Isothermal Amplification’, Clinical Chemistry, 52(2), pp. 303 LP – 306. doi: 10.1373/clinchem.2005.057901.
- Harper, S. J., Ward, L. I. and Clover, G. R. G. (2010) ‘Development of LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosa for quarantine and field applications.’, Phytopathology. United States, 100(12), pp. 1282–1288. doi: 10.1094/PHYTO-06-10-0168.
- Qin, Z.-Q. et al.(2018) ‘Field Evaluation of a Loop-Mediated Isothermal Amplification (LAMP) Platform for the Detection of Schistosoma japonicum Infection in Oncomelania hupensis Snails’, Tropical medicine and infectious disease. MDPI, 3(4), p. 124. doi: 10.3390/tropicalmed3040124.
- Santoiemma, G., Tamburini, G., Sanna, F., Mori, N., & Marini, L. (2019). Landscape composition predicts the distribution of Philaenus spumarius, vector of Xylella fastidiosa, in olive groves. Journal of Pest Science, 92(3), 1101-1109.
- Morente, M., Cornara, D., Moreno, A., & Fereres, A. (2018). Continuous indoor rearing of Philaenus spumarius, the main European vector of Xylella fastidiosa. Journal of applied entomology, 142(9), 901-904.
- Ahern, S. J. et al.(2014) ‘Characterization of novel virulent broad-host-range phages of Xylella fastidiosa and Xanthomonas’, Journal of Bacteriology. American Society for Microbiology Journals, 196(2), pp. 459–471. doi: 10.1128/JB.01080-13.
- Das, M. et al.(2015) ‘Control of Pierce’s Disease by Phage’, PLOS ONE. Edited by A. Rashed. Public Library of Science, 10(6), p. e0128902. doi: 10.1371/journal.pone.0128902.
- Yadeta, K. A. and J Thomma, B. P. H. (2013) ‘The xylem as battleground for plant hosts and vascular wilt pathogens’, Frontiers in plant science. Frontiers Media S.A., 4, p. 97. doi: 10.3389/fpls.2013.00097.
- Bae, C. et al.(2015) ‘Infection processes of xylem-colonizing pathogenic bacteria: possible explanations for the scarcity of qualitative disease resistance genes against them in crops.’, TAG. Theoretical and applied genetics. Theoretische und angewandte Genetik. Germany, 128(7), pp. 1219–1229. doi: 10.1007/s00122-015-2521-1.
- Ionescu, M. et al.(2016) ‘Promiscuous Diffusible Signal Factor Production and Responsiveness of the Xylella fastidiosa Rpf System’, mBio. Edited by C. S. E. Harwood Leo He, Ya-Wen, 7(4), pp. e01054-16. doi: 10.1128/mBio.01054-16.
- Chen, AH. et al. (2015) “Transplantability of a circadian clock to a noncircadian organism”, Science Advances. Vol 1, no. 5, e1500358. Doi: 10.1126/sciadv.1500358.
- Newman, K. L., Almeida, R. P., Purcell, A. H., & Lindow, S. E. (2003). Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera. Appl. Environ. Microbiol., 69(12), 7319-7327.
- Levin, B. R., & Bull, J. J. (2004). Population and evolutionary dynamics of phage therapy. Nature Reviews Microbiology, 2(2), 166.
- RUTBERG, L. (1982). Temperate bacteriophages of Bacillus subtilis. In Bacillus Subtilis (pp. 247-268). Academic Press.
- Gill, J. J., & Hyman, P. (2010). Phage choice, isolation, and preparation for phage therapy. Current pharmaceutical biotechnology, 11(1), 2-14.
- Wagner, P. L., & Waldor, M. K. (2002). Bacteriophage control of bacterial virulence. Infection and immunity, 70(8), 3985-3993.
- Summer, E. J., Enderle, C. J., Ahern, S. J., Gill, J. J., Torres, C. P., Appel, D. N., ... & Gonzalez, C. F. (2010). Genomic and biological analysis of phage Xfas53 and related prophages of Xylella fastidiosa. Journal of bacteriology, 192(1), 179-190.
- World Health Organisation. (2018, February 15). Antimicrobial resistance. Retrieved October 15, 2019, from https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance.
- Chan, B. K., & Abedon, S. T. (2012). Phage therapy pharmacology: phage cocktails. In Advances in applied microbiology (Vol. 78, pp. 1-23). Academic Press.
- Jones, J. D., & Dangl, J. L. (2006). The plant immune system. nature, 444(7117), 323.
- Rapicavoli, J. N., Blanco-Ulate, B., Muszyński, A., Figueroa-Balderas, R., Morales-Cruz, A., Azadi, P., ... & Roper, M. C. (2018). Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nature communications, 9(1), 390.
- Felix, G., Duran, J. D., Volko, S., & Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. The Plant Journal, 18(3), 265-276.
- Denoux, C., Galletti, R., Mammarella, N., Gopalan, S., Werck, D., De Lorenzo, G., ... & Dewdney, J. (2008). Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Molecular plant, 1(3), 423-445.
- Berendsen, R. L., Pieterse, C. M., & Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends in plant science, 17(8), 478-486.
- Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., & Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. New Phytologist, 206(4), 1196-1206.
- Wistrand-Yuen, E., Knopp, M., Hjort, K., Koskiniemi, S., Berg, O. G., & Andersson, D. I. (2018). Evolution of high-level resistance during low-level antibiotic exposure. Nature communications, 9(1), 1599.
- Killiny, N., & Almeida, R. P. (2009). Xylella fastidiosa afimbrial adhesins mediate cell transmission to plants by leafhopper vectors. Appl. Environ. Microbiol., 75(2), 521-528.
- Kim, W. S., & Geider, K. (2000). Characterization of a viral EPS-depolymerase, a potential tool for control of fire blight. Phytopathology, 90(11), 1263-1268.
- Campanharo, J. C., Lemos, M. V. F., & de Macedo Lemos, E. G. (2003). Growth optimization procedures for the phytopathogen Xylella fastidiosa. Current Microbiology, 46(2), 0099-0102.
- Mendes, J. S., Santiago, A. S., Toledo, M. A., Horta, M. A., de Souza, A. A., Tasic, L., & de Souza, A. P. (2016). In vitro determination of extracellular proteins from Xylella fastidiosa. Frontiers in microbiology, 7, 2090.
- Tharanathan, R. N., & Kittur, F. S. (2003). Chitin—the undisputed biomolecule of great potential.
- Nicastro, J., Sheldon, K., & Slavcev, R. A. (2014). Bacteriophage lambda display systems: developments and applications. Applied microbiology and biotechnology, 98(7), 2853-2866.
- Lander, G. C., Evilevitch, A., Jeembaeva, M., Potter, C. S., Carragher, B., & Johnson, J. E. (2008). Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure, 16(9), 1399-1406.
- Pires, D. P., Cleto, S., Sillankorva, S., Azeredo, J., & Lu, T. K. (2016). Genetically engineered phages: a review of advances over the last decade. Microbiol. Mol. Biol. Rev., 80(3), 523-543.
- Gibson, D. G., Benders, G. A., Andrews-Pfannkoch, C., Denisova, E. A., Baden-Tillson, H., Zaveri J., ... & Merryman, C. (2008). Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. science, 319(5867), 1215-1220.
- Garamella, J., Marshall, R., Rustad, M., & Noireaux, V. (2016). The all E. coli TX-TL toolbox 2.0: a platform for cell-free synthetic biology. ACS synthetic biology, 5(4), 344-355.
- C. Bragard et al., “Update of the Scientific Opinion on the risks to plant health posed by Xylella fastidiosa in the EU territory,” EFSA J., vol. 17, no. 5, May 2019.
- Navarro, C., Fernández-Escobar, R., & Benlloch, M. (1992). A low-pressure, trunk-injection method for introducing chemical formulations into olive trees. Journal of the American Society for Horticultural Science, 117(2), 357-360.


