Type 2 Diabetes Mellitus and Incretin Mimetics
Type 2 Diabetes Mellitus (DM) is a metabolic disease affecting approximately 422 million people worldwide [1], while the number is still increasing. One of its major symptoms is the relative lack of insulin, which causes a decreased uptake of glucose into cells, resulting in an energy crisis within the cells. In Type 2 DM, the body compensates this relative lack of insulin by increasing its secretion. This causes stress within the insulin-producing beta cells of the pancreas, eventually leading to their destruction. Consequently, the body is left with an absolute lack of insulin and substitution becomes necessary.
In Type 2 DM, the relative lack of insulin can have multiple reasons. It can be induced by overconsumption of sugar-containing food, decreasing the sensitivity of insulin receptors, as their expression is downregulated. It can also have genetic reasons, such as a mutation in an essential gene.
Incretins, such as glucagon like peptide 1 (GLP-1), are used in the therapy of Type 2 DM. GLP-1 and its analogues are especially interesting for the treatment of Type 2 DM, since they do not only have a direct effect on the pancreas, but also decrease gastrointestinal motility and secretion, hunger, and the emptying of the stomach [2]. Therefore, they help with weight reduction. Additionally, they are considered to be cardioprotective [2]. This is very important for the therapy, since many cases of Type 2 DM are caused by an unhealthy lifestyle and obesity. These effects propose the outlook of using the drug as medication against obesity and adipositas, without the indication of diagnosed Type 2 DM or in stages of prediabetes, in addition to a lifestyle intervention.
GLP-1 and Exendin-4, an analogue of GLP-1, will bind to their receptor on the pancreas surface, which will cause an intracellular cascade activating an Adenylate Cyclase. If ATP is available within the cell, which means if the cell has already taken up sugar and run through glycolysis, cAMP will be produced. Overall, this will result in prolonged depolarization and calcium ion influx and consequently more secretion of insulin from the beta cells [2]. This increase in insulin secretion can overcome the relative lack of insulin in the periphery.

We decided to formulate our drug as a probiotic, since it bears multiple advantages. First of all, a probiotic can implement itself in the human guts microbiome, making it possible that the drug does not have to be taken daily according to a strict therapy scheme. This will also address the group of patients who are not able to stick to a therapy scheme, for instance the elderly or people with fear of other forms of formulation. Also, it was shown by multiple studies that probiotics can, in general, positively influence the course of Diabetes mellitus [3, 4].
Exendin-4, a GLP-1 analogue
At the beginning of our research for drugs for Type 2 Diabetes Mellitus, we were made aware of multiple problems with the usage of human GLP-1, by Dr. Timo Dirk Müller, acting director of the IDO (Institute for Diabetes and Obesity) of the Helmholtz-Institute Munich (Experts). After discussing our first draft of the project, he pointed out several weak points, which we had to work on. First of all, he was concerned about the fast degradation (2 to 5 minutes) by DPP-IV (Dipeptidylpeptidase) and hydrolysis of human GLP-1, which we originally wanted to use as our gene of interest. Consequently, only 5% of GLP-1 reaches the pancreas, which would not be sufficient, if we wanted to have a therapeutic effect. Lastly, Timo explained that the efficiency of the synthesis of our gene of interest must be taken into account, in order to have a therapeutic effect.

Design considerations
We searched for alternatives and set our minds to Exendin-4 [5], since it solely consists of a protein sequence that can be expressed within a bacterium. In humans, Glucagon like peptide 1 is produced in L-cells in the intestines, depending on multiple signaling factors such as nutrients. It originates from the prepeptide proglucagon, which is cleaved into, among others, GLP-1 (1-37) by prohormone convertase 1 (PC1). The inactive GLP-1 (1-37) is then processed into its active form GLP-1 (7-37) by an endopeptidase. This active form is secreted and diffuses across the basal lamina and enters the lamina propria where it is taken up into the capillaries, reaching the circulatory system [2,6]. This natural way of distribution had to be considered in our project, as well.

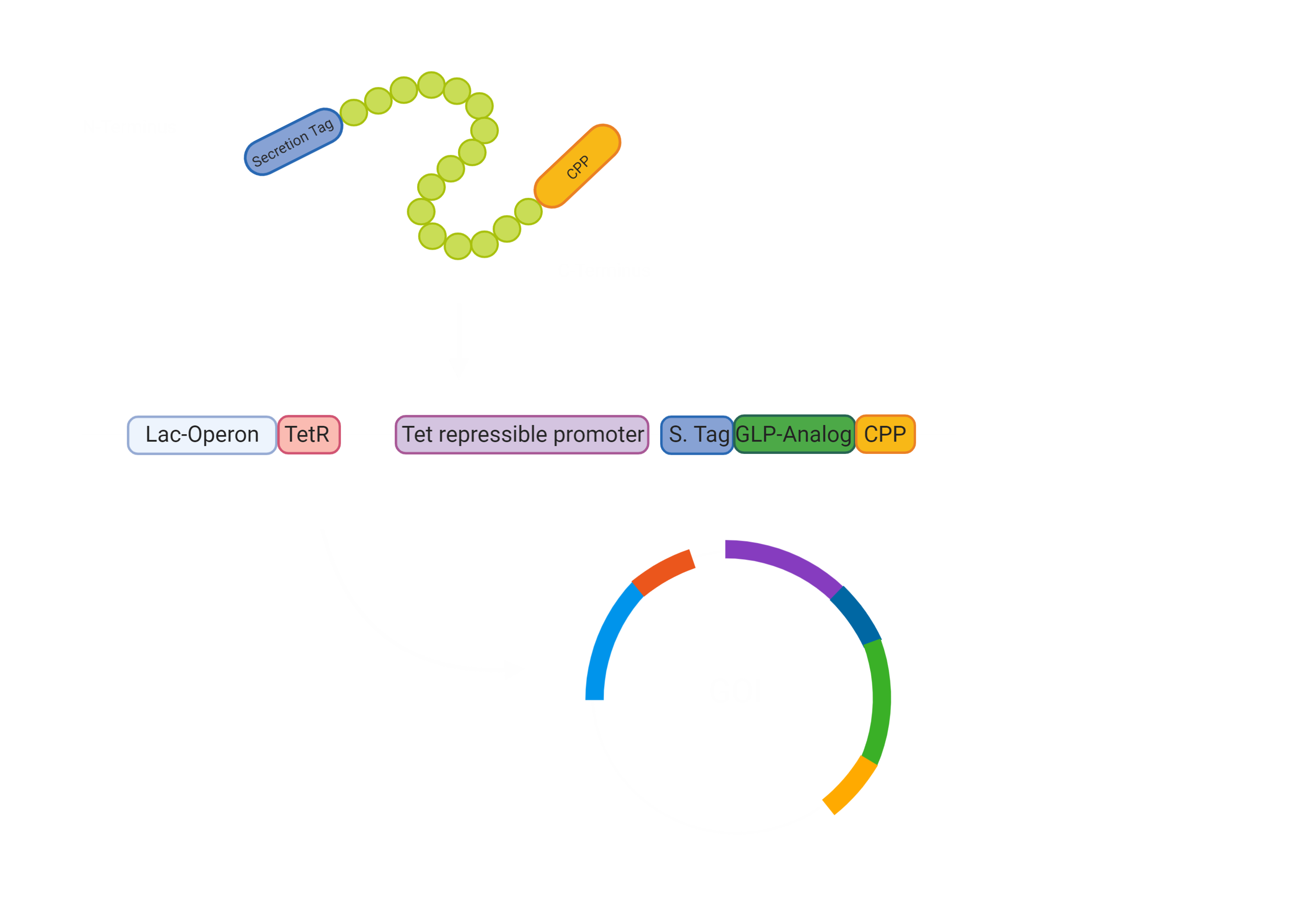
First of all, our drug needs to be secreted by E. coli Nissle [3]. Hence, we made a fusion protein of Exendin-4 with a secretion tag on the N-terminus. After secretion, the protein has to pass the intestinal wall [3]. To achieve this, we decided to fuse a cell penetrating peptide onto the C-terminus of the protein. For the evaluation of a suitable Cell Penetrating Peptide, we also developed a software tool to predict the cargo transport efficiency( Project/Software) In the beginning we were concerned that the use would impair a Biosafety Level 1 usage, however, after consulting with Biosafety representatives, we were sure that the use of Penetratin in our experimental setup will not pose a threat in any way. The biosafety representatives were Brigitte Walderich, who is the representative of the Max Planck Institute Tübingen and Jörg Schibel, the representative of the university clinic Tübingen ( Experts) .

Expression of our gene of interest
Since we could now bring the Exendin-4 to the pancreas, we wanted to make sure there was no unnecessary burden to the pancreas, leading to pancreatitis. Hence, we constructed a glucose dependent secretion based on catabolite repression and the utilization of the Tet repressor. If the glucose concentration in the intestines is low, the bacterial Adenylate Cyclase is active and synthesises cAMP (cyclic AMP), which interacts with CAP (catabolite activating protein). The CAP-cAMP complex then binds to the CAP binding site [7] upstream of the TetR-gene, allowing for the expression of the Tet Repressor TetR, since the transcription of the repressor is activated. The TetR accordingly binds and blocks the activity of the Tet repressible promoter, which controls the expression of our GOI construct.
If the glucose availability increases after the intake of a meal, Adenylate Cyclase is inhibited and the CAP-cAMP complex dissolves [7]. Therefore, the TetR is not active and the GOI can be expressed. To sum up, the presence of glucose in the gut drives the expression of Exendin-4 and therefore increases the release of Insulin only, if food is taken up and Insulin is required.

Final System
Overall, this leaves us with the following construct for the secretion of our Gene of interest. This is encoded on a plasmid and will not be integrated into the genome. This allows us to promote our chassis as an interchangeable system for other drugs, which can just be added through stable transformation.
Laboratory Results
Exendin-4
The first steps were to design our Exendin-4 construct with the N-terminal secretion tag and a C-terminal cell-penetrating peptide (CPP) (BBa_K3096042) . Furthermore, we had to consider how we would combine the Exendin-4 with the Tet repressible promoter and the TetR gene under the control of the lac operon. For the lac operon, we faced some challenges since it contained a lacI binding site that would hinder our planned system to work. We therefore needed to alter the lac promoter by removing the lacI binding site while keeping the rest of the promoter unchanged (BBa_K3096011) .
After the design of the parts and system (BBa_K3096044) was done, we started off with creating and amplifying the two parts of our gene of interest system (Fig.7). On the one hand the lac operon promoter with the TetR gene and on the other hand the Tet Promoter with our modified Exendin-4. The Tet Promoter was pasted in front of the modified Exendin-4 construct via a primer. Our modified Exendin-4 with N-terminal secretion tag and CPP under the control of the Tet promoter was amplified and created via PCR (Fig. 7 second column) and shows the expected size of around 450 bp (BBa_K3096045). The lac operon promoter with the TetR gene (BBa_K3096012) can be seen in the third column in figure 7 with the expected size of around 1 kb.

E. coli cells were successfully transformed with both constructs, however, we could unfortunately not evaluate the basal expression of the Exendin-4 construct without Tet repression since we missed a readout and Exendin-4 specific antibodies were not available or provided to us.
We faced many difficulties when trying to transform and unite both generated parts ( BBa_K3096012 , BBa_K3096045) into one by restriction ligation to our final construct (BBa_K3096044). Especially the ligation process failed several times and we only received backbone religations. We changed the protocol a few times in order to find the right way to ligate our constructs and also tried Gibson Assembly. Even after several troubleshooting meetings with our supervisors we still could not succeed in the ligation of our final expression system (BBa_K3096044).
Cell-penetrating peptides (CPP)
In the context of our Incretin expression system we also focused on the evaluation of different cell-penetrating peptides (CPP) for their ability to enter cells. This is of interest to find the best suitable CPP that has to be fused with our Exendin-4 to allow for the best results and the highest bioavailability in the end. For the evaluation process we designed constructs consisting of a N-terminal His-Tag linked to a GFP, which is fused to a CPP, which can be readily exchanged by restriction ligation. The different CPP parts can be accessed on our Parts subpage.
We further designed a predictive software tool, which allows assigning a transport efficiency score to CPPs. This allows making educated decisions on the design process. For more information on CPPs, the software tool and results please visit our software subpage (Project/Software).
Due to a lack of time we did not manage to create all CPP parts with a His-tag linked to GFP in the laboratory ( BBa_K3096026 , BBa_K3096029 , BBa_K3096035 , BBa_K3096036 , BBa_K3096037 , BBa_K3096038 ).
Next steps
We have successfully designed all the parts which are essential for our glucose-dependent Exendin-4 expression system. The two separate constructs, Glucose-dependent TetR expression (BBa_K3096012) and the modified Exendin-4 under the control of the Tet promoter (BBa_K3096045) can be readily tested for the expression of TetR or Exendin-4 when using a proper readout such as Western Blot.
As a next step, the two parts need to be ligated together, a process into which we invested a lot of time, yet did not succeed. Different incubation times or ligation techniques from the ones we already used will have to be applied (Project/Notebook) .
Once our Incretin expression system is constructed, the glucose-dependent expression of Exendin-4 can be evaluated by supplying different amounts of glucose to the culture medium and analyzing the Exendin-4 levels via western blot or ELISA (enzyme-linked immunosorbent assay).
For our CPP evaluation, we will analyze the efficiency of each of our different CPPs with the GFP-CPP constructs linked to a His-tag. Those constructs will be expressed by bacteria and purified via their His-tag. Then, the purified GFP-CPP constructs will be applied onto human cells, for instance HEK293T cells, and the cell-penetrating capacity evaluated over the GFP signal and fluorescence microscopy. Furthermore, we aim to perform MTT-assays to evaluate the CPPs’ cytotoxicity, a crucial step prior to human application. The most promising CPP will be selected for our final system and linked to Exendin-4.
For proof-of-concept, in the future, we will proceed with experiments on human cell lines. Firstly, the transport of the Exendin-4 through a CaCo-2 cell monolayer will be tested, to investigate, whether the drug can reach the pancreas. Secondly, we will work with Rat Insulinoma cell line INS-1 cells, to examine whether Exendin-4 will induce an insulin expression.
References
- World Health Organization. (2016). Global Report on Diabetes[Accessed 26.03.2019].
- Jens Juul Holst. The Physiology of Glucagon-like Peptide 1. (2007). Physiological Reviews. p.1409-1439.
- Duan F, Curtis KL, March JC. Secretion of insulinotropic proteins by commensal bacteria: rewiring the gut to treat diabetes. Appl Environ Microbiol. (2008);74(23):7437–7438. doi:10.1128/AEM.01019-08
- Duan FF, Liu JH, March JC. Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes. (2015);64(5):1794–1803. doi:10.2337/db14-0635
- Copley, Kathrin & McCowen, Kevin & Hiles, Richard & L Nielsen, Loretta & Young, Andrew & Parkes, David. (2006). Investigation of Exendin-4e Elimination and Its In Vivo and In Vitro Degradation. Current drug metabolism. 7. 367-74. 10.2174/138920006776873490.
- Lim, Gareth E., Brubaker, Patricia L. Glucagon-Like Peptide 1 Secretion by the L-Cell. (2006). 10.2337/db06-S020. Diabetes. p. S70-S77
- Griffiths AJF, Miller JH, Suzuki DT, et al. An Introduction to Genetic Analysis. 7th edition. New York: W. H. Freeman; (2000). Catabolite repression of the lac operon: positive control. Available from: https://www.ncbi.nlm.nih.gov/books/NBK22065/































