08 Jakob Patrick Eva After moving into our new lab and settling in, first practical steps like aliquoting our primers or preparing LB-Medium and LB-Agar were taken. 09 Jakob Patrick Eva We amplified the cas3 gene out of E. coli gDNA by PCR (Q5 Polymerase) and transformed the biobricks BBa_K608351 (K2), BBa_K091001 (K5), BBa_I13453 (K10), BBa_K584000 (AraC Promotor), BBa_K117008 (LsrR Promotor), BBa_R0073 (Mnt Promotor) in competent E. coli DH5-Alpha (NEB iGEM Kit). 10 Eva Patrick We amplified cascade genes out of E. coli MG1655 gDNA via PCR (Q5 Polymerase) and checked the amplification of cas3 and cascade via agarose gel electrophoresis. (Fig. 1) We picked colonies for all but the Ba_K608351 (K2) transformations (no growth) for inoculating overnight cultures and made a new batch of competent E. coli DH5-Alpha cells. 11 Jakob Eva Patrick By Miniprep (Qiagen) we extracted the DNA of the overnight cultures. Amplification via PCR (Q5 Polymerase) of cascade was repeated (Fig. 2). BBa_K608351 (K2) was transformed again and the competent cells tested.


15 Jakob Patrick Eva Luzi Transformations of DH5-Alpha cells with the biobricks were repeated. 16 Patrick, Eva Since there repeatedly weren’t any colonies for BBa_K608351 (K2), we transformed NovaBlue™ competent cells with this biobrick. Overnight cultures of the other biobricks as well as an overnight culture of E. coli Nissle were inoculated. 17 Eva Patrick Katharina Glycerol stocks of the overnight cultures were prepared. Competent E. coli Nissle cells were produced and the growth of E. coli Nissle was analysed preliminarily. After Miniprep (Qiagen) of BBa_K516030 (rfp) a PCR (Pfu Polymerase) was run. After digestion of the PCR products cas3 and cascade as well as the plasmid BBa_I13453 (K10), K10_cascade and K10_cas3 were ligated. 18 Eva Patrick The K10_cascade and K10_cas3 ligations were digested (single) and analysed by gel electrophoresis, together with the rfp PCR product (Fig. 3). The gel did not show a result for K10_cascade and K10_cas3. rfp (PCR Product) and BBa_R0073 (Mnt Promotor) were digested and ligated. 19 Eva Patrick Digestion and Ligation of cascade, cas3 and K10 were repeated with increased amounts of DNA. DH5-Alpha cells were transformed with K10_cascade, K10_cas3 and BBa_R0073(Mnt)_rfp

23 Eva Jakob Patrick Preparation of LB-Medium, LB-Agar and Chloramphenicol plates. Overnight cultures for K10_cascade, K10_cas3 and BBa_R0073(Mnt)_rfp colonies were inoculated. After double digestion of BBa_K584000 (AraC Promoter), BBa_K117008 (LsrR Promotor), the fragments ParaC and PLsrR were extracted from the gel. 24 Eva Patrick After Miniprep of K10_cascade, K10_cas3 and BBa_R0073(Mnt)_rfp (Qiagen) the plasmids were single digested. Gel electrophoresis showed the empty backbone (Fig. 4). The isolated fragments ParaC and PLsrR were ligated into the backbone pSB1K3 after digestion. pSB1K3_ParaC and pSB1K3_PLsrR were transformed in DH5-Alpha cells. 25 Eva Overnight cultures for pSB1K3_ParaC and pSB1K3_PLsrR colonies were inoculated. 26 Eva After Miniprep (Qiagen) of pSB1K3_ParaC and pSB1K3_PLsrR, the plasmids were digested. Gel electrophoresis showed successful ligations (Fig. 5).


30 Patrick, Katharina The team moved into a new laboratory. The synthesized sequences (K0, K3, K4, K6, K7, K15) arrived and were aliquoted. Chloramphenicol plates were prepared. cascade, cas3, BBa_I13453 (K10), rfp and BBa_R0073 (Mnt Promotor) digestions and ligations were repeated and transformed. 31 Patrick Jakob Overnight cultures for K10_cas3 colonies were inoculated, there was no growth of the other transformations. A PCR (Pfu polymerase) was run on the synthesized constructs, it wasn’t successful. The Minipreps of pSB1K3_ParaC and pSB1K3_PLsrR were used for retransformation. 01 Patrick The K10_cas3 overnight culture was miniprepped. Overnight cultures for pSB1K3_ParaC, pSB1K3_PLsrR, as well as K10_cascade and BBa_R0073(Mnt)_rfp (some late colonies grew) were inoculated. 02 Patrick Glycerol stocks for pSB1K3_ParaC were prepared, there was no growth in pSB1K3_PLsrR overnight cultures. After Miniprep of rpSB1K3_ParaC, K10_cascade and BBa_R0073(Mnt)_rfp the plasmids (also K10_cas3) were digested and a gel electrophoresis was run. It showed the transformation of empty backbone in K10_cascade and BBa_R0073(Mnt)_rfp, successful ligation for pSB1K3_ParaC and unsuccessful ligation for K10_cas3 (Fig. 6).

05 Patrick Eva Antonia PCR (Q5 polymerase) and PCR (Paq polymerase) was run on the synthesized constructs, they weren’t successful. cascade and cas3 were successfully amplified via PCR(Q5). 06 Eva Patrick Antonia A gradient PCR (Q5) was run on the constructs. It was discovered that the currently used agarose was falsely prepared with water instead of TAE-Buffer and was discarded. cascade, cas3 and BBa_I13453 (K10) were digested and ligated. 07 Patrick Eva Luzi Antonia A new PCR (Q5) was run on the synthesized constructs, gel electrophoresis suggested positive amplification (Fig. 7). By amplification with accordingly designed primer, K0 has been added a TetR promoter and is now the construct K1. Due to a technical malfunction in the building further work was delayed.

12 Patrick Katharina After respective digestion and DNA clean up, pSBC13_K1, pSBC13_K3, pSBC13_K6, K5_K4, K10_cas3, K10_cascade and BBa_R0073(Mnt)_rfp were ligated. 13 Patrick Eva Antonia Marie DH5-Alpha cells were transformed with Yesterday’s ligations. 14 Luzie Marie Eva A colony PCR (Taq polymerase) was run on pSBC13_K1, but the gel electrophoresis was negative (Fig. 8). Overnight cultures of pSBC13_K1, pSBC13_K3, pSBC13_K6, K5_K4, K10_cas3 and K10_cascade colonies were inoculated. 15 Marie Patrick Eva After Miniprep the plasmids were double digested. Gel electrophoresis suggested successful ligations of pSBC13_K6, K10_cas3, K10_cascade and BBa_R0073(Mnt)_rfp (Fig. 9). 16 Patrick Miniprep of pSBC13_K1 was digested but gel electrophoresis was negative. cascade and cas3 were amplified by PCR (Q5 polymerase). The synthesized construct K2 arrived, was diluted and amplified via PCR (Q5 polymerase).


19 Patrick Jakob Eva After respective digestion and DNA clean up, pSB1C3_K1, pSB1C3_K3, pSB1C3_K7 and K5_K4 were ligated. The BBa_R0073(Mnt)_rfp plasmid was digested to isolate PMnt_rfp by gel extraction (Fig. 10). The backbone pSB1K3 was digested with respective enzymes and pSB1K3_PMnt_rfp was ligated. K1, K2, K3, K4, K7 and K15 were amplified via PCR (Q5 polymerase) with a new programm. 20 Eva Patrick Last week’s pSBC13_K6, K10_cas3 and K10_cascade were sent off to Sanger sequencing. DH5-Alpha cells were transformed with yesterday’s ligations as well as retransformed with K10_cascade . 21 Marie Zoe Patrick A new batch of competent cells was prepared and overnight cultures of pSB1C3_K1, pSB1C3_K7, K5_K4 and K10_cascade inoculated. 22 Patrick Eva Sanger Sequencing was positive on K10_cascade. Chloramphenicol and Kanamycin plates were prepared. After Miniprep of pSB1C3_K1, pSB1C3_K7, K5_K4 and K10_cascade the plasmids were digested and run on a gel. Digestions of pSB1C3_K1 and K5_K4 were negative. The new competent cells were transformed with pSB1C3_K3, pSB1K3_PMnt_rfp and iGEM competent cell test plasmids. 23 Marie Patrick The constructs K1, K2, K3, K4, K7 and K15 were amplified by PCR (Q5 polymerase). Double digestion of pSB1C3_K7 and K10_cascade was repeated. The gel electrophoresis suggested successful ligation of K7, the result for K10_cascade was unclear (Fig. 11).


26 Patrick A new PCR (Q5 polymerase) was run on the constructs and the successfully amplified DNA purified. K10_cascade and pSB1C3_K6 were retransformed. Inoculation of overnight cultures for K10_cascade and K10_cas3 old colonies. 27 Patrick Eva Jakob Antonia Miniprep of K10_cascade and K10_cas3 followed by digestion and gel electrophoresis, which suggested successful ligation of K10_cascade, K10_cas3 is XY (Fig. 12). All synthesized constructs (K0-K15) were digested and ligated with their respective backbone. Overnight cultures of K10_cascade and pSB1C3_K6 retransformations were inoculated. 28 Patrick Eva Antonia The BBa_R0073(Mnt)_rfp plasmid was digested to isolate PMnt_rfp and the backbone pSB1K3 was digested with respective enzymes. pSB1K3_PMnt_rfp was ligated. DH5-Alpha cells were transformed with yesterday’s construct (K0-K15) ligations. Overnight cultures of K10_cascade and pSB1C3_K6 were miniprepped and digested. The gel electrophoresis showed bands suggesting a successful ligation of K10_cascade and pSB1C3_K6 (Fig. 13). The synthesized constructs were amplified via PCR (Q5 polymerase) for our Experimenta collaboration. 29 Eva Antonia Patrick A colony PCR (Q5) was run on K5_K4, the other transformations didn’t show growth. Gel electrophoresis of the PCR didn’t show a positive result. To amplify pSB1C3, a biobrick of the iGEM distribution plates was diluted and a PCR (Q5 polymerase) was run. The constructs (K0-K15) were digested. K0, K2, K3, K15 were ligated with their backbone pSB1C3. K4 was ligated with BBa_K091001 (K5) and K7 was ligated with pSB1C3_K6. The work on the cell penetrating peptides (CPPs) was started by amplifying gfp and synb1 via PCR (Q5 polymerase) (Fig. CPP 1). Primer were dissolved to 100 µM. 30 Antonia Eva Katharina The colony PCR for K4_K5 was repeated, but remained negative. PCR (Q5 polymerase) was run on CPPs. Gel electrophoreses of the PCR showed double bands.


02 Lina Antonia Luzi Jakob Eva Famke As preliminary testing, a growth curve of E. coli Nissle at 37°C, pH 7, was generated. DH5-Alpha cells were transformed with the ligations of constructs (K0-K15) as well as pSB1K3_PMnt_rfp and the distribution kit biobrick. The CPPs tat-lk15, tat, yta2, tp10, pVec and penetratin were amplified by PCR (HotStart Polymerase). The gel electrophoresis showed double bands (Fig. CPP 2). 03/04 Lina Growth curves of E. coli Nissle (EcN) for different temperatures were created. EcN cultures were inoculated with an overnight culture and grown for 120 minutes at 37°C. Next, temperature was changed to 0°C, 4°C, 8°C and 25°C, while one control was kept at 37°C. 37°C and 8°C were chosen for sample preparation for RNA-Seq. EcN was grown 120 min, after being inoculated from one overnight culture, at pH7. Then, media was changed to the respective pH (6, 5, 4, 3, 2, 1) and growth was observed. Growth in pH4 was replicated. EcN was grown in different concentrations of Hydrogen Peroxide (1 µM, 10 µM, 25 µM, 100 µM), infused after two hours of growth at 37°C, pH7 in LB medium. EcN, inoculated from the same overnight culture, were grown under four anaerobic conditions (LB, mGAM anaerobic, mGAM+1.3 mM Metformin, mGAM+0,25mM cholic acid) at 37°C, pH7. EcN aerobic growth in LB was used as control. Samples for RNA sequencing were taken after 270 minutes for all anaerobic conditions, since the increase of pressure within the flasks indicated a change of metabolism of EcN to fermentation processes. Aerobic LB growth samples were taken at OD600=1.0.
EcN, inoculated from the same overnight culture, were grown in mGAM medium under anaerobic conditions at 37°C, pH 7 supplemented with either 50% supernatant of different species or 50% of water as a control for depleted nutrients. Supernatant of Bacteroides thetaiotaomicron, Prevotella copri, Ruminococcus gnavus, Bifidobacterium adolescentis and Clostridium difficile was previously produced by Dr. Lisa Maier. Bacteroides samples were taken for RNA sequencing after 225 minutes. EcN were lastly grown in the growth medium provided by iGEM Stuttgart, in comparison to LB medium. Overnight cultures were used for inoculation of three samples with 5ml each and OD600 was measured every 45 mins. For the dryfreezed samples, 9 µL skim milk were added to 1 µg wet weight of bacteria, frozen at -80°C for 2 h, 42 h at -20°C and 2h at 25°C. Afterwards they were resuspended in LB.
All OD600 measurements were conducted with the Nanospectrometre by Implen. For the results check Project/ Nissle. 04 Eva Antonia A batch of competent cells was prepared. pSB1C3_K0, pSB1C3_K2, pSB1C3_K3, pSB1C3_K15, pSB1C3_K6_K7, K5_K4 and pSB1K3_PMnt_rfp colonies were analyzed by colony PCR(Q5). Gel electrophoresis didn’t show positive result. 05 Eva Antonia Further gel electrophoresis of yesterday’s colony PCR remained negative. Digestions and ligations of constructs were repeated. Overnight cultures of distribution kit biobrick colonies were inoculated. 06 Marie Miniprep of distribution kit biobrick.

10 Eva Antonia K0, K2, K3, K4, K6, K7, K15 were amplified via PCR (Q5 polymerase) using primers designed for Gibson assembly, the PCR didn’t work. After digestion of the CPPs and their backbones, the plasmids pET-28b(+)_synb1 as well as pMiniT2.0_pVec and pMiniT2.0_tat-lk15 were ligated. 11 Eva Antonia A gradient PCR of the constructs was run with Gibson primers, gel electrophoresis suggested successful amplification of K1, K2 and K15 (Fig. 14). 12 Eva Antonia Gibson-PCR of K3, K4, K6, and K7 was successfully repeated. For Gibson assembly the backbones pSB1C3 and BBa_K091001 (K5) were single digested. pSB1C3_K1_K15, pSB1C3_K2_K3, K5_K4 and pSB1C3_K6_K7 were assembled and transformed. Additionally, pET-28b(+)_synb1 was transformed. 13 Eva Antonia The transformations didn’t show colonies so the Gibson assembly was repeated and transformed with provided NovaBlue™ competent cells. Colonies for pET-28b(+) were picked and an over-weekend culture (RT) inoculated. The ligations of pMiniT2.0_pVec and pMiniT2.0_tat-lk15 were transformed in DH5-Alpha cells.

16 Antonia The Novablue™ Gibson transformation showed comprehensive growth so they were diluted by streaking. The over-weekend cultures of pET-28b(+)_synb1 were miniprepped. For pMiniT2.0_pVec and pMiniT2.0_tat-lk15 colonies were picked and overnight cultures inoculated. 17 Marie Eva Antonia The BBa_R0073(Mnt)_rfp plasmid was digested to isolate PMnt_rfp and the backbone pSB1K3 was digested with respective enzymes. pSB1K3_PMnt_rfp was ligated. The dilution streaks of Novablue™ Gibson transformations were diluted again. 18 Marie Eva pSB1K3_PMnt_rfp was transformed. A colony PCR was run on Novablue™ Gibson Assembly colonies. Gel electrophoresis was negative. 19 Marie Antonia The colony PCR was repeated but remained negative. DH5-Alpha cells were transformed with pMiniT2.0_cpp (tat, yta2, tp10, penetratin). A miniprep of pET-28b(+) was performed. The product was controlled by gel electrophoresis but was negative (Fig. CPP 3). 20 Marie Antonia The BBa_R0073(Mnt)_rfp plasmid was digested to isolate PMn_rfp and the backbone pSB1K3 was digested with respective enzymes. pSB1K3_PMnt_rfp was ligated. synb1 was amplified by a PCR. A colony PCR was performed to control the transformed pMiniT2.0_cpp plasmids. Over-weekend cultures (RT) of Novablue™ Gibson Assembly colonies were inoculated.

23 Marie Antonia Transformation of pSB1K3_PMnt_rfp. The over-weekend cultures of Gibson assembly were used for a new colony PCR, lysing the cells before the reaction. It remained negative. pET-28b(+) vector was digested and ligated with synb1. The pET-28b(+)_synb1 was transformed. 24 Marie Eva Overnight cultures of pSB1K3_PMnt_rfp colonies were inoculated. Colony-PCR of pMiniT2.0_cpp was performed, gel electrophoresis showed double bands (Fig. CPP 4). Overnight a ligation of pET-28b(+) and synb1 was performed. 25 Marie Antonia Nanodrop measuring of Gibson-PCR product. Transformation of pET-28b(+)_synb1. After Miniprep of pSB1K3_PMnt_rfp the plasmids were digested. Gel electrophoresis suggested ligation of the empty backbone. Single digestion of pSB1C3 and BBa_K091001 (K5) followed by Gibson assembly was repeated and DH5-Alpha cells transformed. 27 Antonia Marie No growth of new Gibson assembly. Over-weekend (RT) cultures of Novablue™ Gibson Assembly colonies were inoculated. Cultures of pMiniT2.0_penetratin and pET-28b(+)_synb1 colonies were inoculated.

30 Marie Lina Antonia The BBa_R0073(Mnt)_rfp plasmid was digested to isolate PMnt_rfp and the backbone pSB1K3 was digested with respective enzymes. pSB1K3_PMnt_rfp was ligated. Miniprep of Novablue™ Gibson Assembly over-weekend cultures. pSB1C3 was amplified via PCR (Taq polymerase). Transformation of BBa_K592024 (bfp) from the iGEM distribution Kit and inoculation of K2 (BBa_K608351) on plate. Repeating of colony-PCR of pMiniT2.0_penetratin, pMiniT2.0_tat, pMiniT2.0_yta2, pMiniT2.0_tp10 colonies. 01 Marie Lina Antonia Digestion of Gibson assembly Minipreps and gel electrophoresis, which was negative (Fig. 15). Overnight cultures for K2 (BBa_K608351) were inoculated. Newly synthesized cpp+gfp parts arrived. Amplification tp10+gfp, penetratin+gfp, yta2+gfp, pVec+gfp and synb1+gfp by PCR (Fig. CPP 5). 02 Lina Antonia Jakob Miniprep of K2 overnight cultures and digestion. Double digestion of BBa_K091001 (K5) and clean up of the digested backbone. pSB1C3 and the constructs (K0-K15) were amplified via PCR (Taq polymerase). BBa_K575011 (Ev1), BBa_K523001 (Ev2), BBa_K763001 (Ev3), BBa_K763002 (Ev4) and BBa_K398108 (Ev5) from the iGEM distribution kit were transformed in DH5-Alpha cells. Amplification of CPP PM r10 (~100 bp) (Fig. CPP 6). Digest of CPPs. (Fig. CPP6). Retransformation of pET-28b(+). 04 Jakob Lina Antonia Double Digestion of pSB1C3 followed by clean up and Gibson assembly. DH5-Alpha cells were transformed with pSB1C3_K0_K15, pSB1C3_K2_K3, K5_K4 and pSB1C3_K6_K7. Restriction digestion and ligation of pSB1C3_K0, pSB1C3_K2, pSB1C3_K3, K5_K4, pSB1C3_K6, pSB1C3_K7 and pSB1C3_K15 were repeated and transformation performed. Over-weekend (RT) cultures for the distribution kit biobricks were inoculated. Amplification of new CPP : PM penetratin (Fig. CPP 7).




07 Jakob Lina Antonia Overnight cultures of pSB1C3_K0_K15 (Gibson), K5_K4(Gibson), K5_K4 and pSB1C3_K7 were inoculated. Miniprep of Ev1-Ev5 over-weekend cultures. Transformation of BBa_K132018, BBa_K098991, BBa_K876004 and BBa_K74100X from iGEM distribution kit. 08 Antonia Miniprep of construct overnight cultures was conducted, followed by single digest, which suggested unsuccessful ligations. 09 Lina Digest of PM r10 CPP and pET-28b(+) (Fig. CPP 8). 12 Steffen Famke Antonia The characterisation of BBa_K398108 (Ev5), a salt tolerant cluster, was proceeded after identity of the plasmid was confirmed via digestion (Fig. 16). An overnight culture of the bacteria (from which the Ev5 mini originated), as well as DH5alpha wild type cells were inoculated in LB and put on the shaker at 37°C. As a basis for our modeling, the growth of E. coli Nissle 1917 under in minimal media was to be analysed. For this, an overnight culture was inoculated and M9 medium was prepared. 13 Lina Jakob Famke Antonia The salt tolerance experiment was performed. For this LB 0M KCl, 0.05M KCl, 0.1M KCl, 0.2M KCl, 0.4M KCl and 0.8M KCl in LB were prepared and sterile filtered. The experiment was then performed in triplicates with each DH5alpha wild type and the salt cluster bearing DH5alpha cells. Of each 100µL were inoculated in 15 ml of the respective medium. OD600 was measured every 45min using the Nanophotometer of Implen. The experiment was finished upon reaching the end of the exponential phase. The measured data (Fig. 17), was logarithmized, nonlinearly fitted and normalised against each other (Fig. 18), yielding the result that the salt tolerance cluster does not confer to increased tolerance against KCl in E. coli DH5alpha. The minimal medium M9 was inoculated with EcN overnight cultures. Since calcium chloride did not dissolve properly, one group of four samples in M9 was used with the unsolved. After several hours of stirring, another group of M9 with solved calcium chloride was ready. Thus, in total for each medium, 4 samples with 20 ml medium of either LB (control), M9_unsolved and M9_solved were inoculated with 500 µL of EcN and OD600 was measured every 45 min using the Nanophotometer of Implen. The measuring was stopped upon the LB control entering the plateau phase (Fig. 19).
Please find the recipe for the M9 medium provided by Arie Geerlof - Helmholtz Center Munich under the following link: https://static.igem.org/mediawiki/2019/2/20/T--Tuebingen--M9_recipe.pdf

RNA-Seq
For the preparation of the cultures of E. coli Nissle 1917 Lina used LB-Lennox medium. For one sample the pH was adjusted to pH 4 and was incubated at 37°C. In another sample 100 µM H2O2 were added and incubated at 37°C. The remaining three samples were incubated at different temperatures (8,25,37°C).
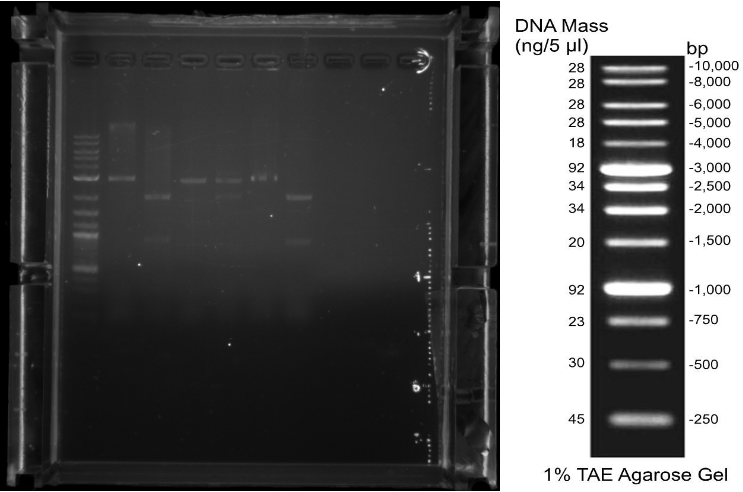
After pelleting the bacterial cultures the pellet was taken up in 1 ml RNAlater to ensure that the RNA is not degraded and expression levels do not change. Patrick and Lina isolated the RNA from the 24 samples with the PureLink RNA Mini Kit by Thermo Fisher Scientific using 500ml of the diluted pellet. The isolated nucleic acid was checked via an agarose gel and the amount estimated via Qubit measurement before being treated with DNase I by Roche.

Patrick purified the RNA via the RNA Clean & Concentrator Kit by Zymo. Another agarose gel was conducted and RNA amount estimated via Qubit. rRNA depletion was conducted with the riboPOOL kit by siTOOLs, the RNA was again purified with the RNA Clean & Concentrator Kit by Zymo and RNA amount estimated via Qubit measurement. Despite very low RNA amount it was proceeded with the Library preparation with the CORALL Total RNA-Seq Library Prep Kit by Lexogen. RNA amounts were analysed via Qubit measurement and Library quality was analysed via FemtoPulse measurement by Agilent. Additionally a qPCR of a pool of all 24 libraries was conducted to better determine the Library amount with the NEBNext Library Quant Kit by NEB. Since the library quality was not sufficient, the rest of the DNase I treated samples was used to start depletion and library preparation again. This time the Total RNA Seq Library Prep Kit by Zymo was used, RNA amount estimated via Qubit measurement and Library quality analysed via FemtoPulse as well as a qPCR. Again RNA amount was low after depletion and library preparation and library quality was not high enough for sequencing. Jakob repeated the E. coli growth and the RNA Extraction with the Qiagen RNeasy kit instead of the dry freezed samples the condition was changed to LB aerob with pH8. The nucleic acid concentration was high after extraction and the RNA showed no sign of degradation.
A DNAse digestion was conducted, the cDNA synthesis, the library preparation and the rRNA depletion were done with the Universal Prokaryotic RNA-Seq Prokaryotic AnyDeplete ™ kit from NuGEN. At a first glance, the library preparation seemed successful. Soon, the pooled libraries will be loaded onto the Illumina MiSeq System for sequencing.
Lina and Katharina prepared the anaerob bacterial cultures with the different conditions: LB aerob, Lb anaerob, mGAM medium anaerob, mGAM medium with 1.3 mM Metformin anaerob, mGAM medium with 0.25 mM cholic acid anaerob and mGAM medium with the supernatant of a Bacteroides thataiotaomicron culture. Cultures were again pelleted and taken up in RNAlater. Patrick isolated the RNA with the PureLink RNA Mini Kit by Thermo Fisher Scientific and conducted a Qubit measurement before and after treatment with DNaseI by Roche. Jakob and Steffen tested the quality of the RNA via an Agilent RNA 6000 Nano Bioanalyzer. Degradation was clearly visible but the samples could be used for the library prep. The rRNA was depleted via the NEBNext (R) rRNA Depletion Kit. Thereafter, our collaborators at the EMBL conducted the automated RNA library preparation and sequencing.