Inspiration
Our team directly addresses the challenge of applying a series of innovative technologies to the development of a practical, affordable and easy-to-use platform for the detection of water-related diseases in developing areas.
The Problem of Cholera
The Problem of current Sensing Methods
Why Aptamers?
Their nature, DNA or RNA, gives them a set of extremely useful characteristics
They are capable of resisting higher temperatures than other molecules, as well as extreme levels of pH or other harsh conditions
They are among the most cost-effective molecules to produce at this point in time
They are among the easiest molecules to manage in a wet lab
Dive into our project







Disease selection & marker expression
After this, this synthetic biological platform (named Escherichia cholira in our proof of concept) went on to the next step: the RoboSELEX.
Keep going to learn more about the building of this platform!
RoboSELEX
Our goal for this year has been the automation of the whole aptamer-selection process (or SELEX) in order to both increase its reproducibility and facilitate the scaling of the pathogenic agents that it is able to detect.
In our vision “Robots making aptamers” we imagine a room full of pipetting robots working full-time constantly discovering new aptamers, so we can scale up our detection system and sense more and more diseases.
Dive into this part of the project under the RoboSELEX section in “Our Technology”!
Characterization
To dig into the qualities of the aptamers and obtain necessary variables such as the affinity constant, we developed several automatic characterization protocols.
Discover how we did this under the “Aptamer Characterization” section.
3D Folding & Computational improvement
Determining structural characteristics as the region where an aptamer and its ligand match can help us to direct further improvements in the most accurate way. This simplifies the SELEX process, as it stops being random, and improves efficacy.
In the course of this year, we took a number of folding algorithms from previous teams and rewrote the folding process. We did this by applying cutting-edge AI techniques for algorithm development, which created a faster and more efficient folding.
Do you want to know more about this process? Have a look at the “Aptamer Folding” section!
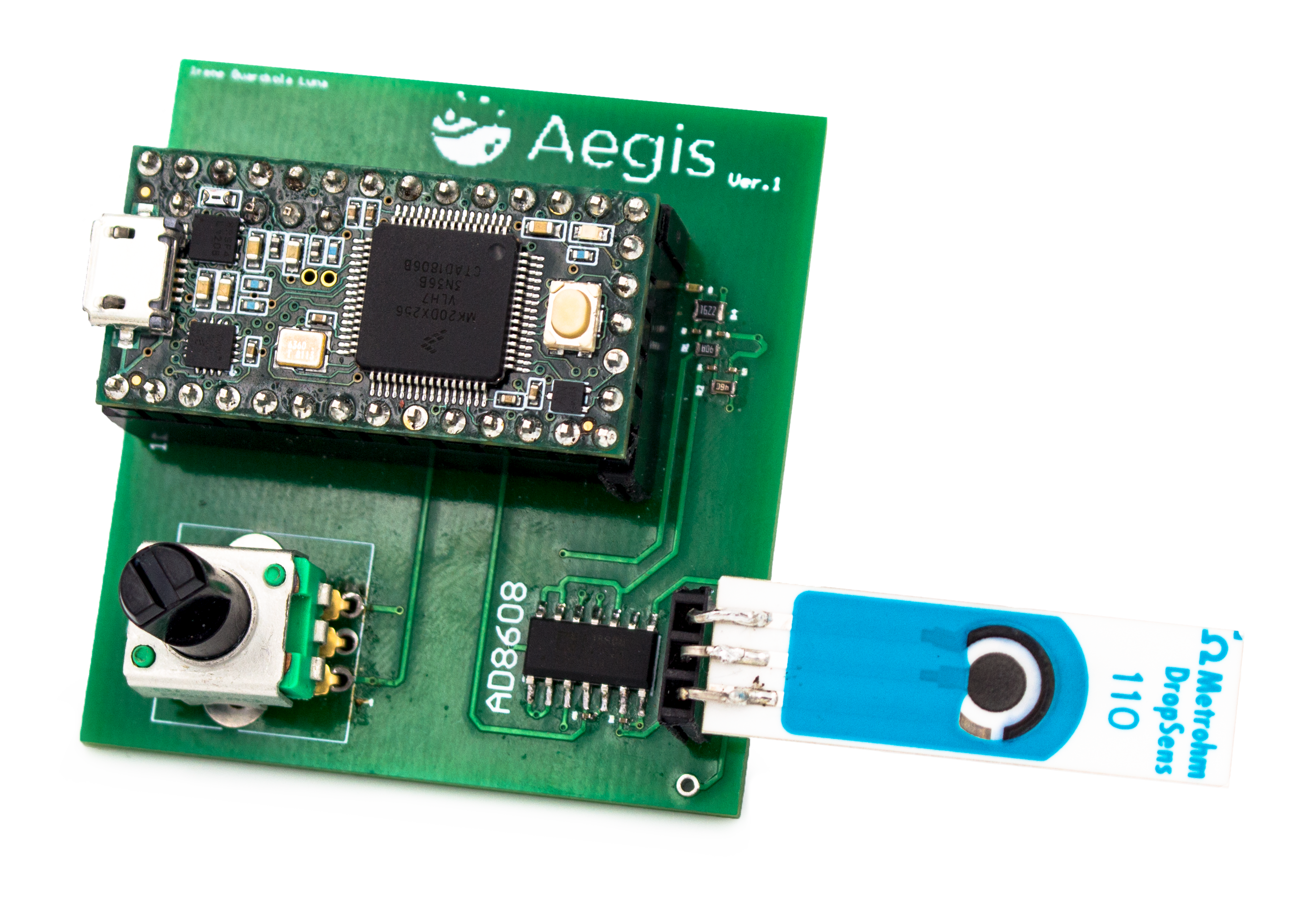
Sensor design & implementation
In this way, the development of the sensor has been intimately linked with our Human Practices, making this entanglement one of the main features of our project. We have based our work on the tangible effect it has on real human lives, by ensuring an interdisciplinary approach that accounts for all aspects of the subject at hand.
To this end, we made our fieldwork in Yaoundé, Cameroon a central part of the project; there, we worked together with local scientists and health workers to tackle the problem of cholera directly, rather than just withdrawing to the lab.
We believe it is essential to not limit science to Western hegemonic knowledge, which recreates colonial patterns of thought, and this community-based fieldwork has allowed us to situate our work amidst - not against - local forms of knowledge. As such, those affected by cholera are just as much part of the project as we are, and will in the future be able to use our protocols in an independent, self-sufficient way.
The sensors
In this way, the development of the sensor has been intimately linked with our Human Practices, making this entanglement one of the main features of our project. We have based our work on the tangible effect it has on real human lives, by ensuring an interdisciplinary approach that accounts for all aspects of the subject at hand.
To this end, we made our fieldwork in Yaoundé, Cameroon a central part of the project; there, we worked together with local scientists and health workers to tackle the problem of cholera directly, rather than just withdrawing to the lab.
We believe it is essential to not limit science to Western hegemonic knowledge, which recreates colonial patterns of thought, and this community-based fieldwork has allowed us to situate our work amidst - not against - local forms of knowledge. As such, those affected by cholera are just as much part of the project as we are, and will in the future be able to use our protocols in an independent, self-sufficient way.




