Project Inspiration and Description
Abstract Project Description
The project of the iGEM Team Tübingen 2019 is the development of a microbial chassis, which can be used as a probiotic E.Coli Nissle 1917, that secretes a drug against Type 2 Diabetes Mellitus. The microbial chassis will consist of a CRISPR/Cas3 based kill switch system, which senses the environmental conditions it is exposed to, consequently determining whether the survival, and integrity of the nucleic acids, of the bacterium can be provided. This kill switch system should enable the use of a genetically engineered organism (GMO) in therapy, as the threat of release into the environment is reduced, since it cannot survive outside the human gut. Thus, it is a concept that can be used for a diverse spectrum of therapies by exchanging the drug and the conditions.
The gene of interest we are focusing on, Exendin-4, serves as a drug for Type 2 Diabetes Mellitus and is an incretin mimetic originating from a lizard called Gila monster (Heloderma suspectum). This incretin mimetic is already used in the pharmaceutical industry for the therapy of Type 2 Diabetes Mellitus and is also known as Exenatid. The human analogue to Exendin-4 is glucagon like peptide 1 (GLP-1), which is secreted from the L-cells in the small intestine, if nutrients are available in the gut. GLP-1 and its analogues cause the so-called incretin effect, which means that the release of insulin as response to sugar is higher, when sugars are consumed orally in contrast to when they are given intravenous.
Project Inspiration
When committing to iGEM, we decided we wanted to a) design a project which uses a new system of the field of synthetic biology and b) make an impact with a product, which can actually be of use to the society. So when Dr. Pengfei Xia, one of our instructors proposed to design a new kill-switch system for bacteria, based on the Type I CRISPR system of CRISPR/Cas3, we were determined to not only implement the system, but also find a way of making it a useful, universally applicable, tool.
As there are four students from the field of molecular medicine within our team, we rapidly got stuck on brainstorming in the field of medical applications for our Kill-Switch systems, whether there was a disease which could easily profit from a new therapeutic strategy. This led us to Diabetes Mellitus, one of the main diseases responsible for premature death worldwide [16]. In 2014, there were 422 Million adults with a diagnosed Diabetes Mellitus worldwide [1], an alarmingly high number, which inspired us to look into alternative treatment strategies of Type 2 Diabetes Mellitus patients, as they make up the largest group of Diabetes patients. There are currently many options for treatment available, however all of these treatments require the patient to actively participate and comply to his or her therapy scheme. As compliance and persistence are key factors for therapeutic success, we, the iGEM Team Tübingen, want to revolutionize the treatment and its administration by using Synthetic Biology and our CRISPR/Cas3 system.
In order to do so, we thought about changes in therapy, which would make it easier to comply to the therapy scheme and easily came up with issues about daily injections and multiple drugs, that need to be taken at certain times a day. Overall, we came to the conclusion that a probiotic bacterium, that synthesises a drug when it is needed and therefore lifts the burden of application from the patient, was our way to go. On top of that, we realized that in 2017 Team AQA_Unesp had already tried to target Type 1 Diabetes Mellitus with a probiotic, hence we felt that the viability of the idea was supported by this [7]. Additionally, this approach would allow us to not only use our bacterium with the CRISPR/Cas3 system as biofactory, but also make it the therapeutic agent which is applied, making the biosafety function of CRISPR/Cas3, not only a tool, but also a requirement for safe application.
The idea of using GMOs as probiotics, is, in general, very interesting for chronic diseases. Therefore, we want to use the application in Type 2 Diabetes Mellitus as an example of the strengths and limitations of such a system, and use our public involvement to gather the society’s perception and opinion of such a therapeutic strategy. Consequently, we set ourselves some goals for the project, which are i) the proof of principle of the CRISPR/Cas3 system ii) the production of Exendin-4 and iii) the incorporation of the public and several sub-projects for communication and education into our project.
Overall, we consider our project to be an important trial for the use of synthetic biology in long-term therapy, where a patient is not restricted to stay within a facility, but can live normally and without a financial or other burden of his or her disease, as the probiotic bacteria in the microbiome will do their job for the medication. We hope, to have designed a safe system which will prohibit the survival of GMOs within the environment and enables the use of probiotics.
Project Description
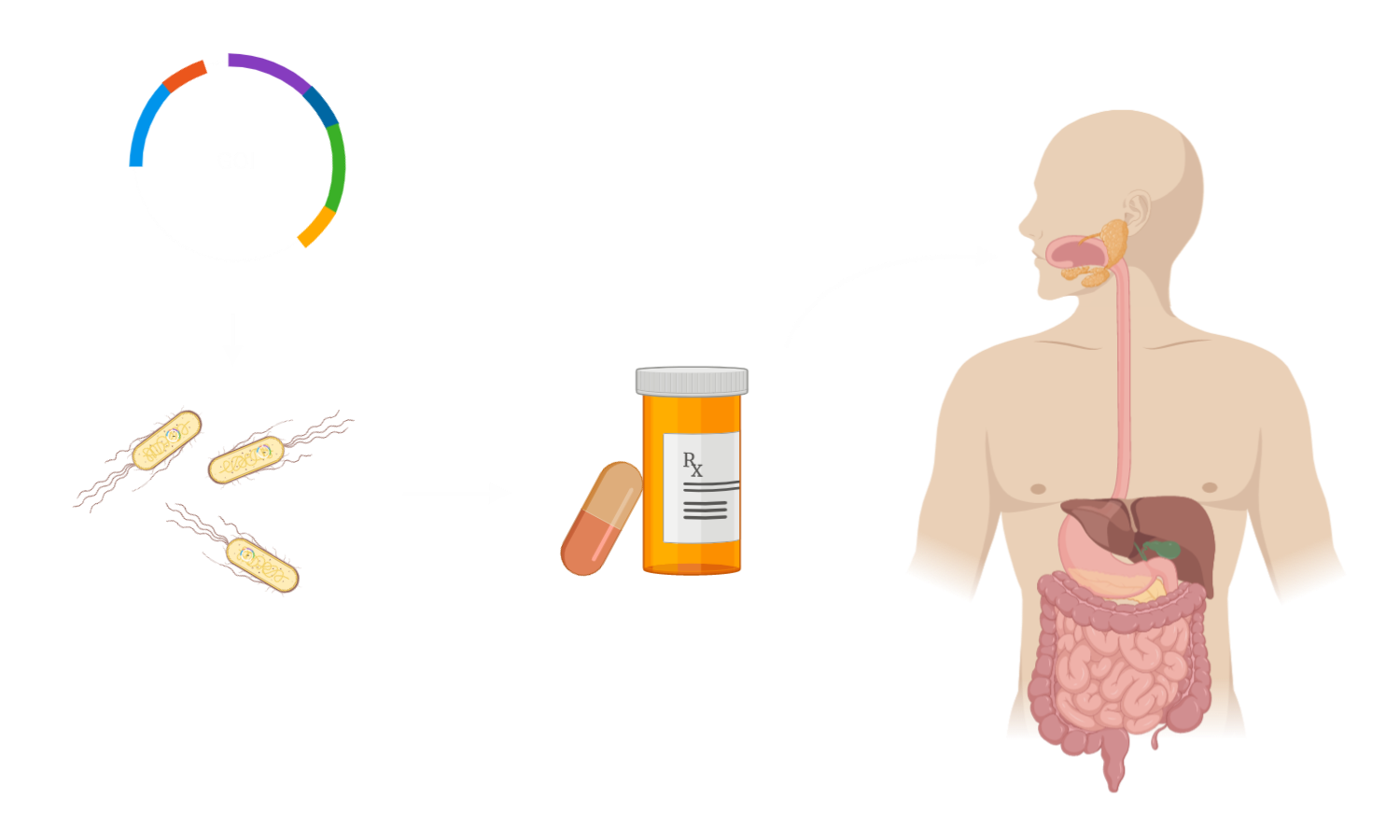
Type 2 Diabetes Mellitus and Incretin Mimetics
GLP-1 and Exendin-4 will bind to their receptor, a G-protein coupled receptor on the pancreas surface, which will cause an intracellular cascade activating an Adenylate Cyclase. If ATP is available within the cell, which means if the cell has already taken up sugar and run through glycolysis, cAMP will be produced. Overall, this will result in prolonged depolarization and calcium ion influx and consequently more secretion of insulin from the beta cell [2]. This increase in insulin secretion can overcome the relative lack of insulin in the periphery, which occurs in Type 2 Diabetes Mellitus.
GLP-1 and its analogues are especially interesting for the treatment of Type 2 DM, since they do not only have a direct effect on the pancreas, but also decrease gastrointestinal motility and secretion, hunger and the emptying of the stomach [2], thus helping in weight reduction. Additionally, it is considered to be cardioprotective [2]. This is very important for the therapy, since many cases of Type 2 DM are caused by an unhealthy lifestyle and obesity. On top of that, this effect proposes the outlook of using the drug as medication against obesity and adipositas, without the indication of diagnosed type 2 DM or in stages of prediabetes, in addition to a lifestyle intervention.
Generally, Type 2 Diabetes Mellitus is a metabolic disease, which affects about 10% of the population worldwide, with increasing numbers. One of its major symptoms is the relative lack of insulin, which causes decreased uptake of glucose into cells, resulting in an energy crisis within the cells. In Type 2 DM, the body compensates this relative lack of Insulin by secreting more Insulin, which causes stress for insulin-producing beta cells of the pancreas. This increased stress will lead to the destruction of the beta cells, leaving the body with an absolute lack of Insulin, making insulin substitution necessary.
In Type 2 DM the relative lack of Insulin can have multiple reasons. It can be induced by overconsumption of sugar-containing food, decreasing the sensitivity of the Insulin receptors, as they are downregulated. But it can also have genetic reasons, like a mutation in an essential gene.

We decided to formulate our drug as a probiotic, since it bears multiple advantages. First of all, a probiotic can implement itself in the human guts microbiome, making it possible that the drug does not have to be taken daily according to a strict therapy scheme. This will also address the group of patients, who are not able to stick to a therapy scheme, for instance the elderly or people with fear of other forms of formulation. Also, it was shown by multiple studies, that probiotics can, in general, positively influence the course of Diabetes mellitus [5,6].
As a consequence, our probiotic can be released into the environment due to excretion. For biosafety reasons and for strict control of our therapy, we wanted to implement a new biocontainment chassis, based on a CRISPR/ Cas 3 system.
CRISPR/Cas3-system

The system is new in its form and was kindly provided to us by Dr. Pengfei Xia, based on---. Under conditions, which are common in a healthy human beings intestines, 37°C, availability of fatty acids in form of Acyl CoA and N-Acetyl-Glucosamin (GlcNAc), released by the metabolism of mucus through commensal microorganisms [9], our chassis allows for the existence of a plasmid, which carries our gene of interest (GOI) and the survival of the bacterium.
Consequently, if the conditions are according to the ones in the intestine, the Cas3 protein and casABCDE genes cannot be expressed and the CRISPR arrays for the self-targeting of the plasmid and the genome are not transcribed. However, if the environmental conditions change, for instance because the bacterium is excreted, the Cas3 system will be activated. Finally, this will end in the degradation of the foreign plasmid and the genomic DNA, killing the bacterium.
We chose the Cas3 system, because unlike Cas9, who is involved in CRISPR/Cas system II, Cas3 is a component of the CRISPR/Cas sytem type I. Type I systems consists of several proteins but do also target double-stranded DNA. The major advantage of Cas3 compared to Cas9 in our approach is the fact that Cas3, when activated, degrades the whole nucleic acid. The incorporation of the Cas3 system into our probiotic therefore, ensures that once the probiotic leaves its designated environment the Cas3 system degrades all genetic information. By using a targeting array for the bacterium’s genome and one for the plasmid with the gene of interest a spread of nucleic acid is prevented allowing for a safe therapy. Our chassis with the CRISPR/Cas3 system can also serve as a foundation for other applications since it creates a biosafety probiotic that can be modified by exchanging the gene of interest without loosing its unique safety standards.
The regulation of our kill switch is based on three double negated sets of biosensors combined with repression systems.

A) Temperature sensing with a permissible temperature of 37°C
A constitutively active promoter expresses Clts, a temperature sensitive cI repressor of the lambda phage. If the temperature is significantly below 37°C (body temperature), Clts is an active repressor for the cI lambda promoter. This promoter controls the transcription of a repressor protein, AraC. Therefore, if conditions are not 37°C, the AraC is not available, hence the pBAD promoter, controlled by AraC, is active and Cas3 and CasABCDE are expressed, which allows for the Kill-Switch induction. Accordingly, if the bacterium is within the body and the temperature is at 37°C, Clts is unstable and cannot repress the expression of AraC. AraC consequently inhibits the pBAD promoter and the enzymes for the self-kill are not available, the bacterium survives. The usage of the Clts and following FadR sensing system was the iGEM NTU Taida Project of 2012 [8].
B) Fatty acid availability inhibiting the plasmid self targeting array
The intake of a fatty meal increases the fatty acid availability within the body and therefore their metabolite Acyl-CoA will increase. Long chain Acyl-CoA will bind constitutively expressed FadR and hence inhibits its activity [12,13]. FadR is the repressor of the promoter pFad, which regulates the expression of the LsrR repressor, which itself represses the the pLsrR promoter [14] regulating the transcription of the plasmid self targeting array. If the bacterium leaves the body, there will be no fatty acids (Acyl-CoA) available. This allows the activity of FadR, which inhibits pFad. Thus, there will be no LsrR and the self targeting array is active. If the self targeting array of the plasmid is active, the Cas3 complex can use it to target the plasmid and degrade it, inducing the degradation of the foreign DNA.
C) N-Acetyl-Glucosamin-6 Phosphate of metabolized Mucin inhibits the genomic self-targeting array
Commensal bacteria in the gut metabolize the mucus within the intestines, which increases the level of GlcNAc within the lumen [9]. GlcNAc is taken up by the bacteria through their PTS system and metabolized into GlcNAc-6-P, which binds the repressor protein nagC. If nagC is bound to GlcNAc-6-P it loses its abilities to bind DNA, and therefore its respective regulation activity. In our case, nagC can consequently not serve as a repressor of the nag Operon nagBACDE, anymore [10, 11]. Thus, a repressor protein, the Mnt repressor of the Lambda phage, can be expressed. The Mnt repressor inhibits the transcription of the genomic self targeting array, which is controlled by the Mnt promoter. The genomic DNA cannot be degraded.
Nonetheless, if the bacterium leaves the body and the human microbiome, its GlcNAc-6-P sources will deplete and the nagC repressor remains active, repressing the expression of the MntR and lastly, allowing the transcription of the CRISPR array and the Cas3-targeting of the genome, which will kill the bacterium.
Exendin-4, a GLP-1 analogue
At the beginning of our research for drugs for Type 2 Diabetes Mellitus, we were made aware of multiple problems with the usage of human GLP-1, by Dr. Timo Dirk Müller, acting director of the IDO (Institute for Diabetes and Obesity) of the Helmholtz-Institute Munich. After discussing our first draft of the project, he pointed out several weak points, which we had to work on. First of all, he was concerned about the fast degradation (2 to 5 minutes) by DPP-IV (Dipeptidylpeptidase) and hydrolysis of human GLP-1, which we originally wanted to use as gene of interest. Consequently, only 5% of GLP-1 reaches the pancreas, which would not be sufficient, if we wanted to have a therapeutic effect. Lastly, Timo explained that the efficiently of the synthesis of our Gene of Interest must be taken into account, in order to have a therapeutic effect.

Thus, we searched for alternatives and set our minds to Exendin-4 [4], since it solely consists of a protein sequence that can be expressed within a bacterium. In the human, Glucagon like peptide 1 is produced in L-cells in the intestines, depending on multiple signalling factors, such as nutrients. It originates from the prepeptide proglucagon, which is cleaved into, among others, GLP-1 (1-37) by prohormone convertase 1 (PC1). The inactive GLP-1 (1-37) is then processed into its active form GLP-1 (7-37) by an endopeptidase. This active form is secreted and diffuses across the basal lamina and enters the lamina propria, where it is taken up into the capillaries, reaching the circulatory system [2,3]. This natural way of distribution had to be considered in our project, as well.

First of all, our drug needs to be secreted by E. Coli Nissle [5]. Hence, we made a fusion protein of Exendin-4 with a secretion tag on the N-terminus. After secretion, the protein has to pass the intestinal wall [5]. To achieve this, we fused a cell penetrating peptide,onto the C-terminus of the protein. For the evaluation of a suitable Cell Penetrating Peptide, we researched multiple options and finally decided on Penetratin, which is derived from Drosophila Antennapedia Homeodomain [22]. In the beginning we were concerned that the use would impair a Biosafety Level 1 usage, however, after consulting with Biosafety representatives, we were sure that the use of Penetratin in our experimental setup will not pose a threat in any way. The biosafety representatives were Brigitte Walderich, who is the representative of the Max Planck Institute Tübingen and Jörg Schibel, the representative of the university clinic Tübingen.

Since we could now bring the Exendin-4 to the pancreas, we wanted to make sure there was no unnecessary burden to the pancreas, leading to pancreatitis. Hence, we constructed a glucose dependent secretion based on Catabolite repression and the utilization of the Tet repressor. If the glucose concentration in the intestines is low, the bacterial Adenylate Cyclase is active and synthesises cAMP (cyclic AMP), which interacts with CAP (catabolite activating protein). The CAP-cAMP complex then binds the CAP binding site [15] upstream of the TetR-gene, allowing for the expression of the Tet Repressor TetR, since the transcription of the repressor is activated. The TetR accordingly binds and blocks the activity of the Tet repressible promoter, which controls the expression of our GOI construct.
If the glucose availability increases after the intake of a meal, Adenylate Cyclase is inhibited and the CAP-cAMP complex dissolves [15]. Therefore, the TetR is not active and the GOI can be expressed. To sum up, the presence of glucose in the gut drives the expression of Exendin-4 and therefore increases the release of Insulin only, if food is taken up and Insulin is required.

Overall, this leaves us with the following construct for the secretion of our Gene of interest. This is encoded on a plasmid and will not be integrated into the genome. This allows us to promote our chassis as an interchangeable system for other drugs, which can just be added through stable transformation.
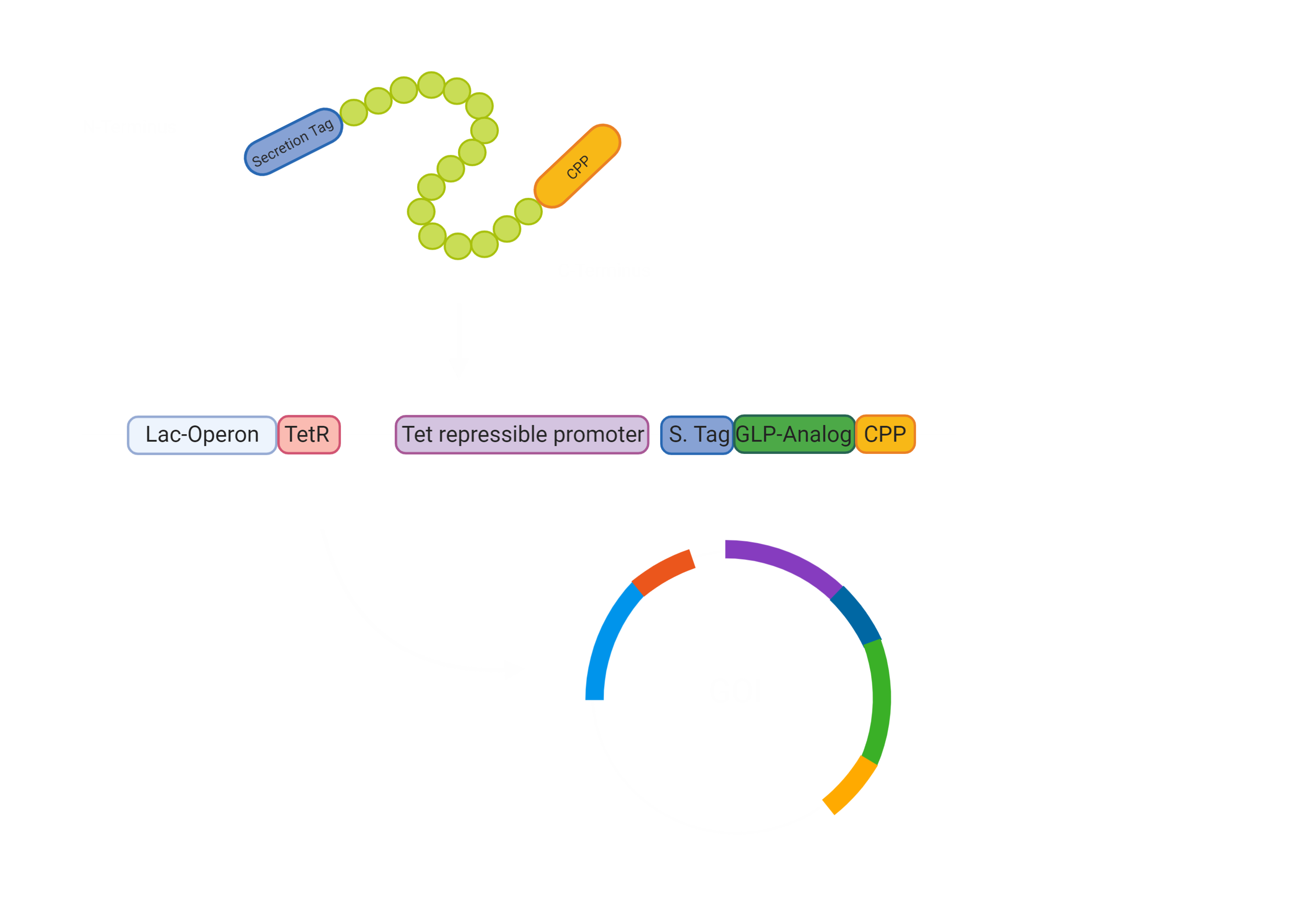
Wetlab Project Plan
To conclude, our projects will require intensive cloning of multiple regulatory elements. For the parts, we will use Biobricks that were sent with the 2019 shipping, Biobrick sequences from the database of iGEM, as well as new sequences, which are unique to our project, like the Exendin-4 fusion construct or the Glc-NAc-6-P sensing system. After synthesis, the GOI constructs need to be amplified and cloned into complete constructs, which will be used to transform competent E. Coli NEB10 cells, as they allow for easier manipulation. Here, we can already test whether our GOI construct work and Exendin-4 is secreted in a Glucose dependent manner.
At the same time the Cas3 system will be isolated from E. Coli K12, and the DNA with regulatory system will be changed in E. Coli K12 to our individual sensing mechanisms. The functionality of the kill switch and the different conditions for it can be tested already. If the construct shows the desired activity, it will be integrated into the E. Coli Nissle 1917 genome. In the end, we hope to combine our GOI with the Cas3 positive E. Coli Nissle 1917 and test the functionality of our system as a whole. Hence, if E. Coli Nissle 1917 secretes Exendin-4 in a Glucose dependent manner and if it kills itself, if the conditions of the gut are not met.
If the system works, we will proceed with human cell line experiments. First of all, the transport of the Exendin-4 through a CaCo-2 cell monolayer to test, whether there will be a drug, which can reach the pancreas. Secondly, we will work with Rat Insulinoma cell line INS-1 cells, to check whether Exendin-4 will induce an Insulin expression.
Modelling and Simulation
Drylab
While our biocontainment strategy relies on the Class I Type I CRISPR system, we will utilize CRISPR/Cas9 technology for the construction of the chassis. CRISPR/Cas9 (or similar) technology has become a versatile toolkit for gene editing in all domains of life. Despite of the successes, off-target activities can be observed in many cases. These off-target activities lead to unwanted genome alterations when non-homologous end joining machineries are available in the system of interest, and therefore have to be considered in every experiment. To facilitate the analysis of experimental off-target identification, we will create a data analysis pipeline for GUIDE-Seq and CIRCLE-Seq experiments to identify off-target sites in CRISPR/Cas9 experiments. Our goal is to generate a reproducible and parallelizable analysis software tool, based on the bioinformatics workflow management system Nextflow. To us, creating an easy-to-use software tool is of highest priority. Thus, we are creating a clear and comprehensible graphical user interface. We plan to improve the existing workflow layout by replacing single components to improve the accuracy of the result, and to reduce the computational time needed. Furthermore, we will validate the results of our workflow experimentally, and will integrate the results of our software tool into our wet-lab experiments. We will encourage other iGEM teams and research groups, who are working with CRISPR/Cas9 systems, to use our tool.
Human Practice
While checking the scientific possibilities to produce a therapeutic probiotic, we soon realized that we would have to engage the public with the goal of designing a product and Human Practice had to be an important part of our project.
Therefore, we divided our Human Practices project into four phases:
Phase I: General Considerations
Phase II: Human Practice Events of the iGEM Team Tübingen
Phase III: Implementing new strategies based on our public integration and outreach
Phase IV: Conclusion and Reflection
Our Human Practice projects therefore are:
Biosafety measures
Obviously, biosafety is very important if a GMO is used as a probiotic. That’s why, on the one hand we have invested a lot of energy into the design of our CRISPR/Cas3 kill switch system, and on the other hand have intensively confronted ourselves with the iGEM regularies and the german law. As a support, Jörg Schibel and Brigitte Walderich helped us evaluate the safety for parts, we weren’t quite sure whether they were Biosafety Level 1 conform, like the CPP.
Political involvement
Since a GMO as a probiotic is currently not allowed to be used in Germany or the European Union, we are interested in whether politics is moving in this field and what has to be done to achieve change. Hence, we are currently planning a so-called “Fishbowl”, which is a discussion round with one seat that can be taken by anyone from the audience. We want to invite people from the group of Young Europeans, who belong to different parties, and discuss Synthetic Biology, the dynamic of the European Law etc. with them, in order to grasp where we need to take action to promote change, which allows a more diverse usage of Synthetic Biology outside the lab.
Social impact and public reach out
From the beginning, we wanted to choose a project that matters not only to us but can be helpful for the public. Since, Diabetes is now considered a pandemic with a substantial threat to human health around the world and currently, at least 400 Million people worldwide suffering from Diabetes [16], we are sure to have chosen a topic, which requires more attention. Especially, with Germany as a comparatively small country, being currently on rank 9 of total diabetes cases, with 7.5 Million patients [16].
This pandemic is a result of an unhealthy and sedentary lifestyle, which promotes obesity and lack of overall fitness, resulting in not only a health, but a socioeconomic crisis as well. That’s why we want to start a preventive campaign for Diabetes and Adipositas in cooperation with Dr. Eike Latz, Institute Director of the Institute of Innate Immunity Bonn and his partner Dr. Anette Christ. Additionally, we have met with Prof. Andreas Fritsche, the deputy director of the Institute for Diabetes Research and Metabolic Disease of the Helmholtz Institute Munich at the Eberhard-Karls University Tuebingen, who has advised us to set another focus on solely adipose people, since our drug has the potential to help them lose weight. Through Prof. Fritsche, we also got insight into the clinical aspects and patient’s acceptance of our product. Overall, Prof. Fritsche made an estimation, that about two thirds of his Type II DM patients would probably be willing to try a new therapy, such as ours. However, he is concerned that the general belief that genetically modified organisms are dangerous and bad, may pose a threat to our idea.
Consequently, we decided to launch a survey [19]. To design our survey, we wanted to fulfill the criteria proposed by iGEM. Hence, informed consent and privacy/ data protection are highly important in survey conduction. Also, our survey design had to be specific and unambiguous, with the goal of not compromising our survey by insinuating a “correct” response to any question. For this purpose, we used the tools provided for free for students by www.umfrageonline.com. As in our case, the participants volunteered to conduct our survey, our survey is a “nonprobability” survey and the results do not represent a randomized group. However, our study had the purpose to help us with refining our project and give us insight into the perception of new therapies and Synthetic Biology. We launched the survey when one of our teammates explained the project at a science slam, which gave us a great audience and allowed for data collection. However, the students present were not really a group of high Diabetes Mellitus prevalence, therefore, we also asked several Diabetologists in Tübingen to spread the survey amongst their patients. The data is currently being evaluated, so we can draw our conclusions to the project.
Lastly, Prof. Fritsche proposed us to hold a talk about our project in September 2019 for the Diabetes patients, who have taken part in his studies. We are looking forward to presenting the results of our research and getting their opinion on our project.
iGEMxSDGs challenge in collaboration with iGEM TAS_Taipei and iGEM Costa Rica
The Sustainable Development Goals (SDGs) of the United Nations (UN) aim to improve the world’s situation by promoting prosperity, equality, peace and care for the environment. Altogether, the UN has set 17 of these goals, which need to be achieved by 2030 [18]. During a meet-up in germany, a member of the iGEM Team Costa Rica has approached us with their interest to promote the SDGs, as after all iGEM projects generally aim to improve people’s lives, while having great exposure to their communities. When iGEM TAS_Taipei then launched their promotion for their video online conference about the SDGs, we decided to join forces and start a collaboration, called iGEMxSDGs.
iGEMxSDGs is a challenge which invites all teams of iGEM to match their project to the respective SDGs targeted by their teams. Afterwards, the team will post and promote their match on their social media channels, spreading the SDG challenge within the iGEM community. This will, on the one hand, promote the SDGs within the iGEM community and the public, and on the other hand highlight the importance of the SDGs to the teams themselves. Also, iGEMxSDGs will inspire new projects within iGEM, as it raises awareness to global issues which need to be solved.
As the SDGs want to introduce sustainable development, we need to make sure that our efforts are sustainable as well. That’s why we do not only want this challenge to be part of iGEM 2019, but want to let it enter the following years’ competitions, as well. To allow for this sustainability, we have designed a subpage on iGEM Costa Rica’s Wiki, which is not only an information platform, but also a database of the efforts each team went into, to target the SDGs. After all, this gives each iGEM team the opportunity to grasp the impact of the iGEM competition and their work and how each team can make a difference [20].
To introduce the SDGs further, iGEM Team Taipei will host an online video conference in early september, with a discussion centered around the SDGs, as part of a MUN (Model United Nations) project. This will also give iGEM teams the opportunity to align their projects to the SDGs and cooperate with other teams targeting the same aspects [21].
For our own matching, we chose the following goals:




Good health and well being is our first goal, because we want to revolutionize the treatment of Type 2 DM, as well as Adipositas, which is one of the major causes of DM. This includes the reduction of premature mortality caused by non-communicable disease, by promoting the usage of synthetic biology for new treatments.
The second goal would be No poverty, because with our project we aim to lift the financial burden of treatment for type 2 Diabetes Mellitus, by providing a less cost intensive alternative to current treatments. This is also very important, since the numbers of Diabetes cases in the developing countries are massively increasing [16] and the current therapy scheme would not be a viable option for economically weak individuals.
Next, we chose Industry, innovation and infrastructure, since we want to promote innovation within therapy schemes and synthetic biology. Our product could have a big impact on the pharmaceutical industry, allowing GMOs to further enter the industrial market.
As a fourth goal, we chose partnerships, as collaboration is what makes a project successful. We thrive to form partnerships, not only for knowledge and material transfer in the lab, but also for interaction with communities. We are happy to have formed an alliance with iGEM Team Taipei and Costa Rica for the #iGEMxSDGs challenge to promote the SDGs.
Education
We believe that education in the field of Synthetic Biology is key in order to promote it in the society and get rid of the stigma of the “bad” GMOs. Therefore, one of our team members visited her former school and taught the students about our project. The positive feedback inspired us to look further. Therefore, we contacted the Experimenta in Heilbronn, Germany, which is Germany’s biggest Science Centre [17].
Sources
[1] World Health Organization. (2016). Global Report on Diabetes. Available online https://apps.who.int/iris/bitstream/handle/10665/204871/9789241565257_eng.pdf;jsessionid=8D5E18D2A5627102F238B7D2DA7AD2C4?sequence=1 [Accessed 26.03.2019].
[2] Jens Juul Holst. The Physiology of Glucagon-like Peptide 1. (2007). Physiological Reviews. p.1409-1439. https://www.physiology.org/doi/abs/10.1152/physrev.00034.2006
[3] Lim, Gareth E., Brubaker, Patricia L. Glucagon-Like Peptide 1 Secretion by the L-Cell. (2006). 10.2337/db06-S020. Diabetes. p. S70-S77
[4] Copley, Kathrin & McCowen, Kevin & Hiles, Richard & L Nielsen, Loretta & Young, Andrew & Parkes, David. (2006). Investigation of Exenatide Elimination and Its In Vivo and In Vitro Degradation. Current drug metabolism. 7. 367-74. 10.2174/138920006776873490.
[5] Duan F, Curtis KL, March JC. Secretion of insulinotropic proteins by commensal bacteria: rewiring the gut to treat diabetes. Appl Environ Microbiol. (2008);74(23):7437–7438. doi:10.1128/AEM.01019-08
[6] Duan FF, Liu JH, March JC. Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes. (2015);64(5):1794–1803. doi:10.2337/db14-0635
[7] https://2017.igem.org/Team:AQA_Unesp
[8] https://2012.igem.org/Team:NTU-Taida/Project/Circuit
[9] Sicard JF, Le Bihan G, Vogeleer P, Jacques M, Harel J. Interactions of Intestinal Bacteria with Components of the Intestinal Mucus. Front Cell Infect Microbiol. (2017);7:387. Published 2017 Sep 5. doi:10.3389/fcimb.2017.00387
[10] Barnhart MM, Lynem J, Chapman MR. GlcNAc-6P levels modulate the expression of Curli fibers by Escherichia coli. J Bacteriol. (2006);188(14):5212–5219. doi:10.1128/JB.00234-06
[11] Konopka JB. N-acetylglucosamine (GlcNAc) functions in cell signaling. Scientifica (Cairo). (2012);2012:489208. doi:10.6064/2012/489208
[12] https://www.uniprot.org/uniprot/P0A8V6 accessed: 19 Jun 2019
[13] Feng Y, Cronan JE. Crosstalk of Escherichia coli FadR with global regulators in expression of fatty acid transport genes. PLoS One. (2012);7(9):e46275. doi:10.1371/journal.pone.0046275
[14] Federle MJ. Autoinducer-2-based chemical communication in bacteria: complexities of interspecies signaling. Contrib Microbiol. (2009);16:18–32. doi:10.1159/000219371
[15] Griffiths AJF, Miller JH, Suzuki DT, et al. An Introduction to Genetic Analysis. 7th edition. New York: W. H. Freeman; (2000). Catabolite repression of the lac operon: positive control. Available from: https://www.ncbi.nlm.nih.gov/books/NBK22065/
[16] IDF (International Diabetes Federation. IDF Diabetes Atlas. (2017). Eighth Edition. Available from: https://diabetesatlas.org/resources/2017-atlas.html
[17] https://www.experimenta.science/
[18] https://www.un.org/sustainabledevelopment/ accessed: 01 May 2019 logos retrieved from here according to their guidelines
[19] https://igem-tuebingen.com/human_practices survey available under the link
[20] https://2019.igem.org/Team:Costa_Rica/SDG-Challenge iGEMxSDGs page
[21] https://forms.gle/xtoeMHuAMpDHeaCc9 Team Taipei video conference
[22] https://www.jenabioscience.com/probes-epigenetics/cell-labeling/protein-and-nucleic-acid-internalization/single-cell-penetrating-peptides-cpps/cpp-p01-penetratin

