Notebook
Week 3
Week 4
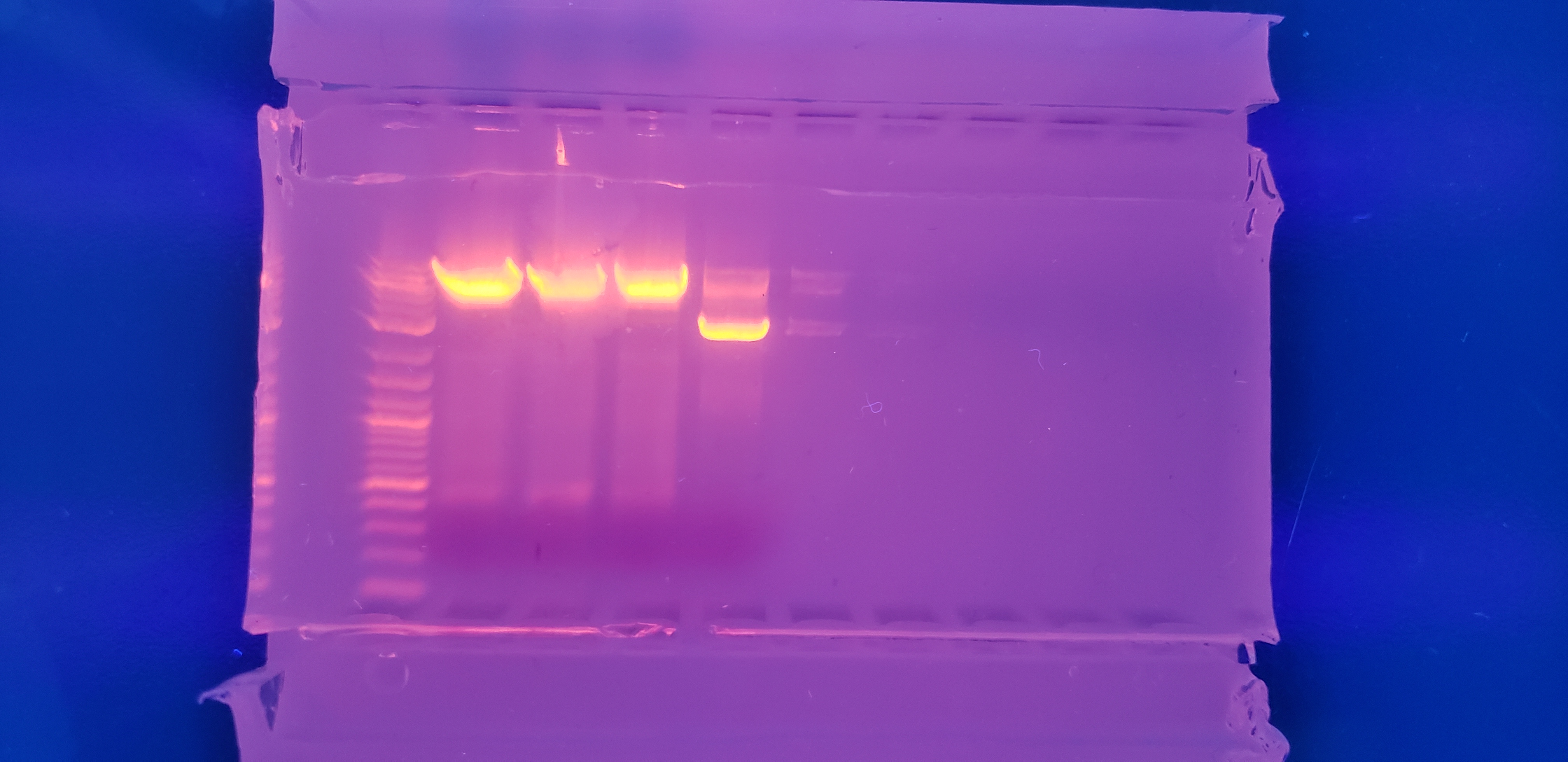
Week 6
Colony PCR - 1
Top: pVS28:(3,4,5,6,7,8,1), pVS29:(1,2,3,4), pVS43:(1,2,3,4), pVS29:(5)
Bottom: pVS29:(6), pVS30:(1,2,3,4,5,6), pVS31:(1,2,3,4,5,6,7,8)
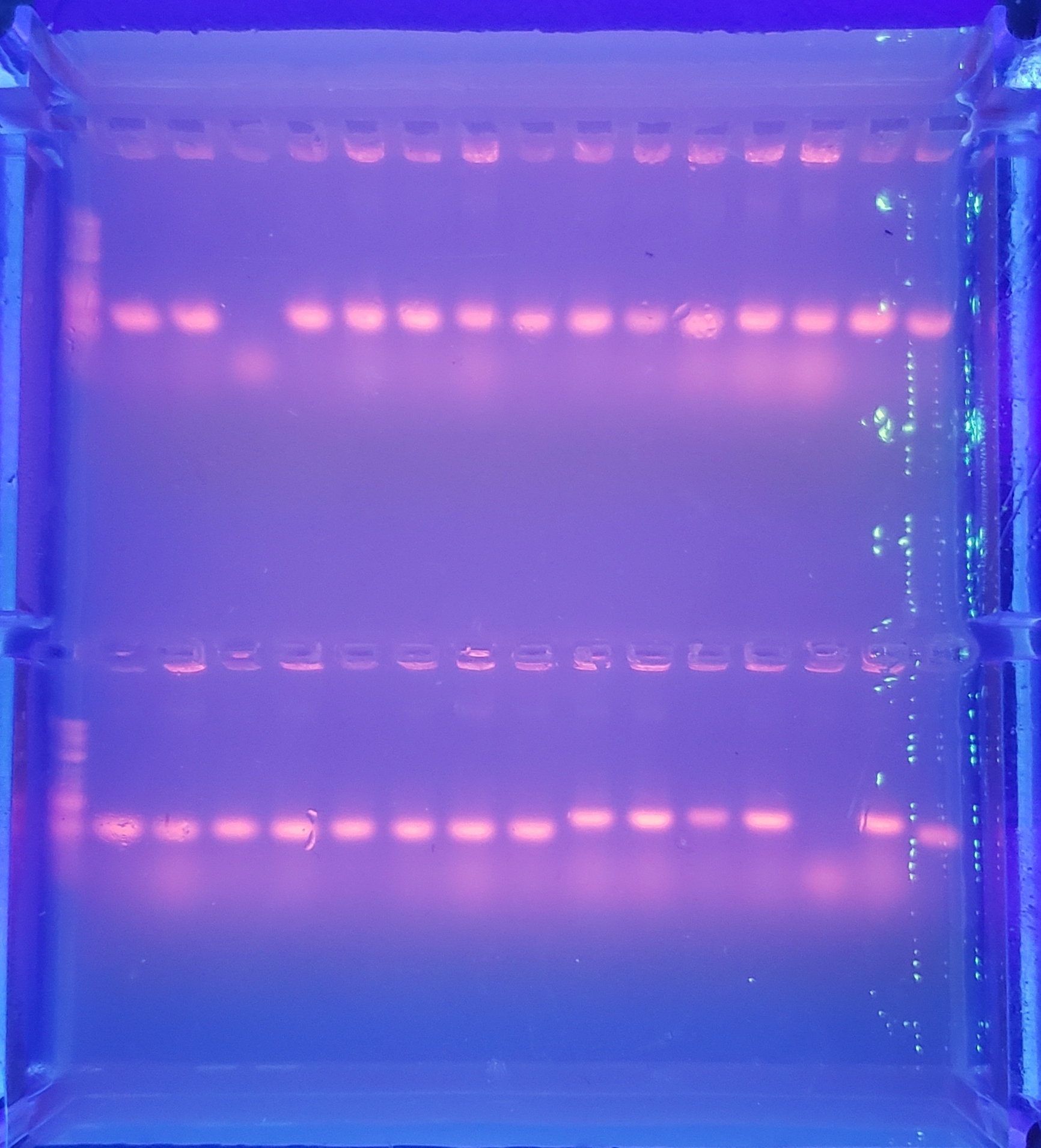
Colony PCR - 2
Top: pVS34:(1,2,3,4,5,6,7,8), pVS33:(1)
Bottom: pVS33:(2,3,4,5,6,7,8), pVS28:(1,2)
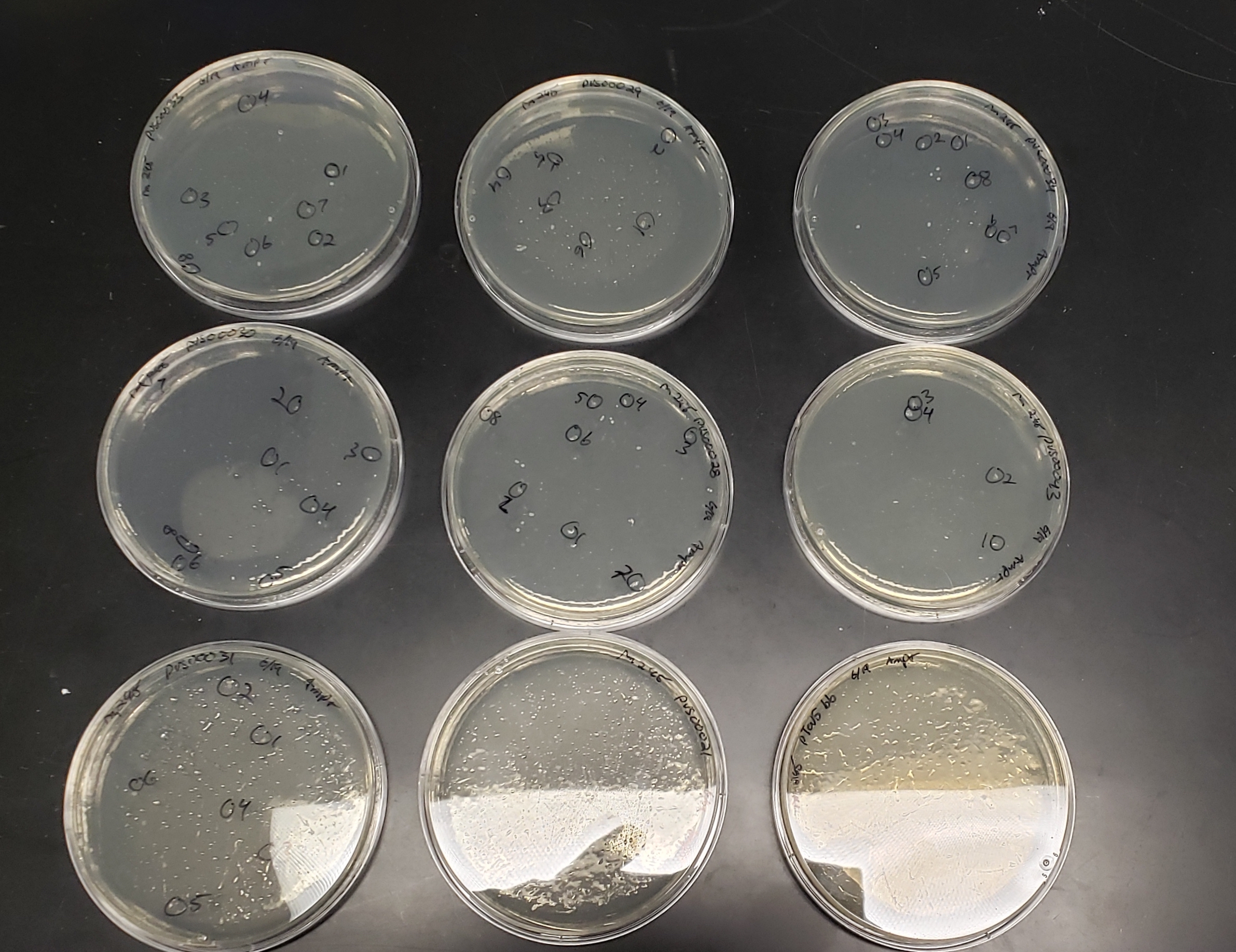
BL21 transformation plates
(slightly overgrown)
Week 7
Expression gel (SDS-PAGE)
Left to right: pVS(28,29,30,31,33,34,43)
Our ladder was old and did not work
(we got better at running gels I promise)
Purification gel (SDS-PAGE)
Insufficient lysis caused protein to be retained in the pellet
Week 8
We tried several different cycle patterns on the sonic dismembrator to determine the best mode of lysis moving forward.
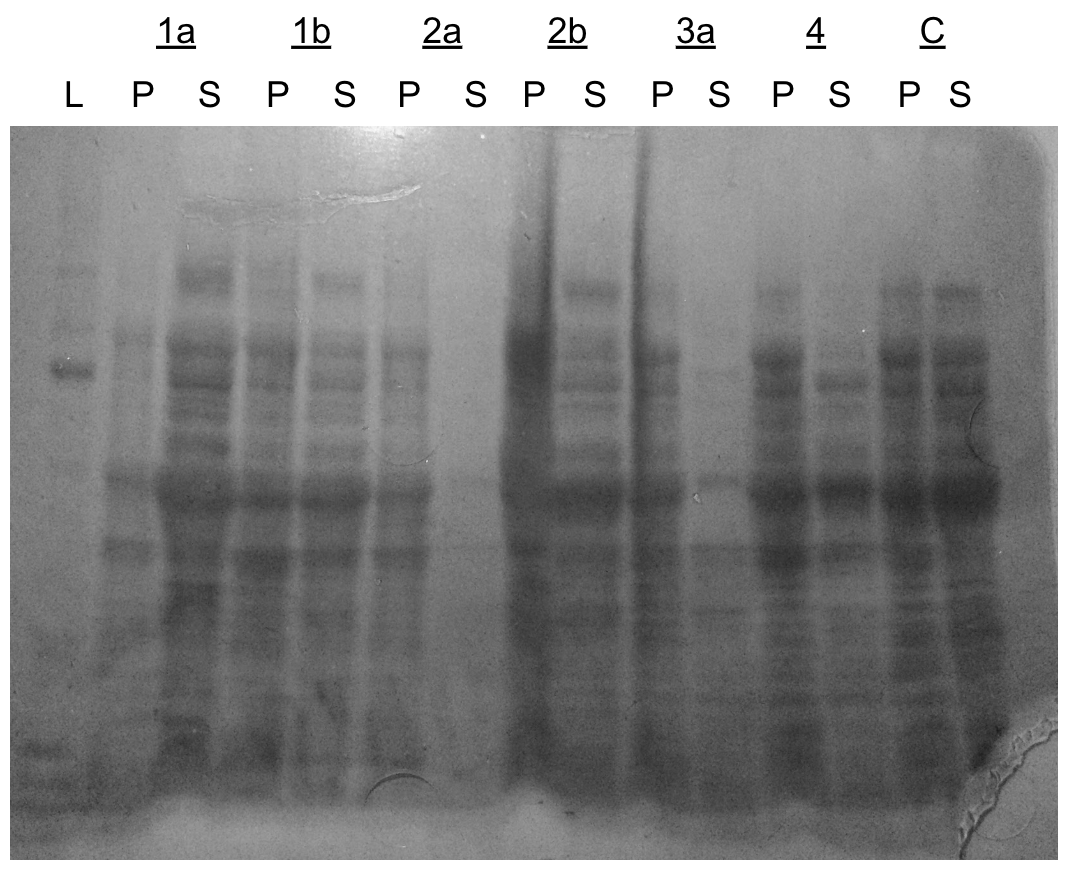
Dismembrator gel (SDS-PAGE)
L=ladder, P=pellet, S=supernatant
- 1a - (10 sec on/20 sec off) x 20
- 1b - (10 sec on/20 sec off) x 40
- 2a - (15 sec on/15 sec off) x 20
- 2b - (15 sec on/15 sec off) x 40
- 3a - (25 sec on/5 sec off) x 15
- 4 - (1.5 min on)
- C - no lysis
Week 9
We attempted another small scale expression test. Cells were grown to OD600=0.5 and induced to 1mM IPTG for 4 hours at room temperature.
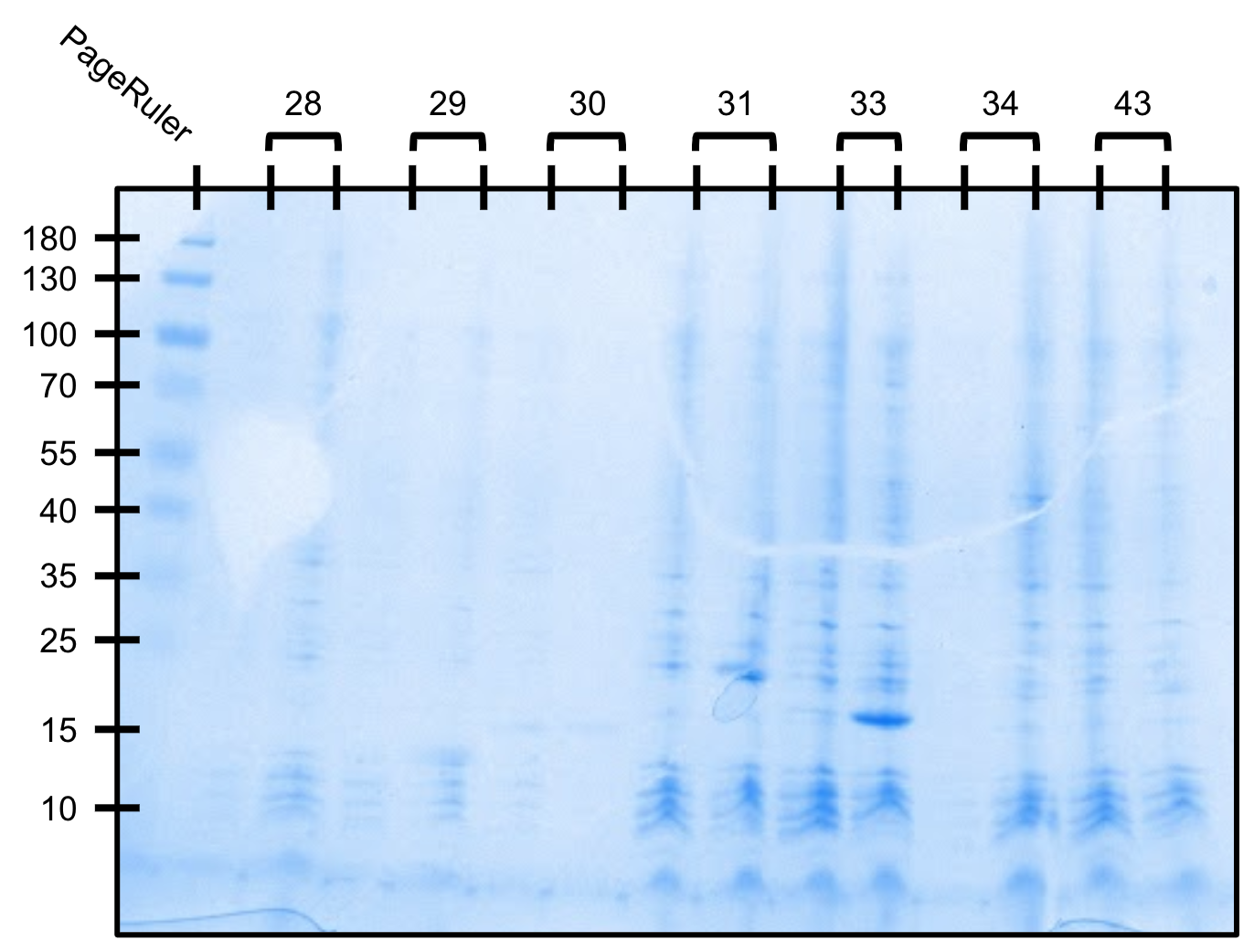
Expression test gel (SDS-PAGE)
For each construct, left=uninduced, right=induced
Starting with a 250mL culture of TEVp, we attempted Ni-NTA purification again using our new lysis protocol. Unfortunately, almost all of the protein was lost in the flow-through indicating poor his-tag binding or a faulty column/protocol.
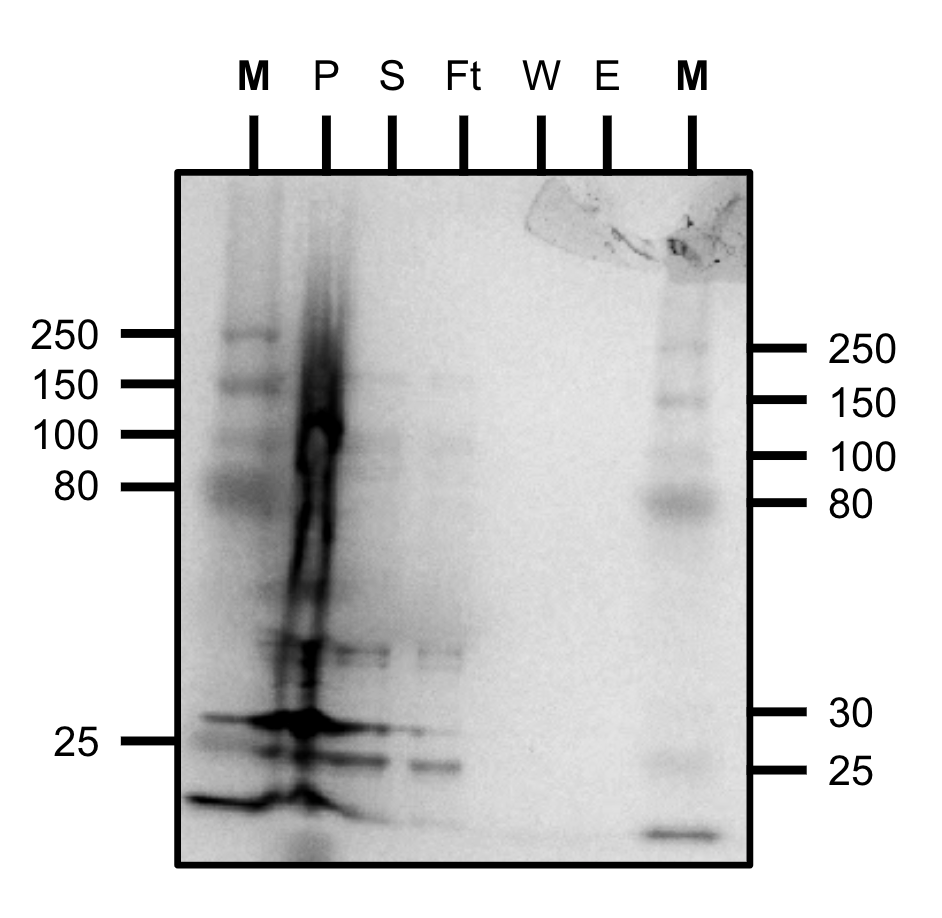
Purification test gel (SDS-PAGE)
M=marker, P=pellet, S=supernatant, Ft=flow-through, W=wash, E=elution
We received cloned-in plasmids from Twist with out set 2 constructs and transformed them into BL21.
We took 20mL cultures of pVS29, pVS31 and pVS33 and attempted Ni-NTA purification. However, based on an A280 reading, we observed no protein in our elution -- likely due to excessive washing.
We expressed 250mL cultures of constructs pVS(29,31,33) for further purification tests.
The lower band for each construct is our protein of interest and the top band is LacI.
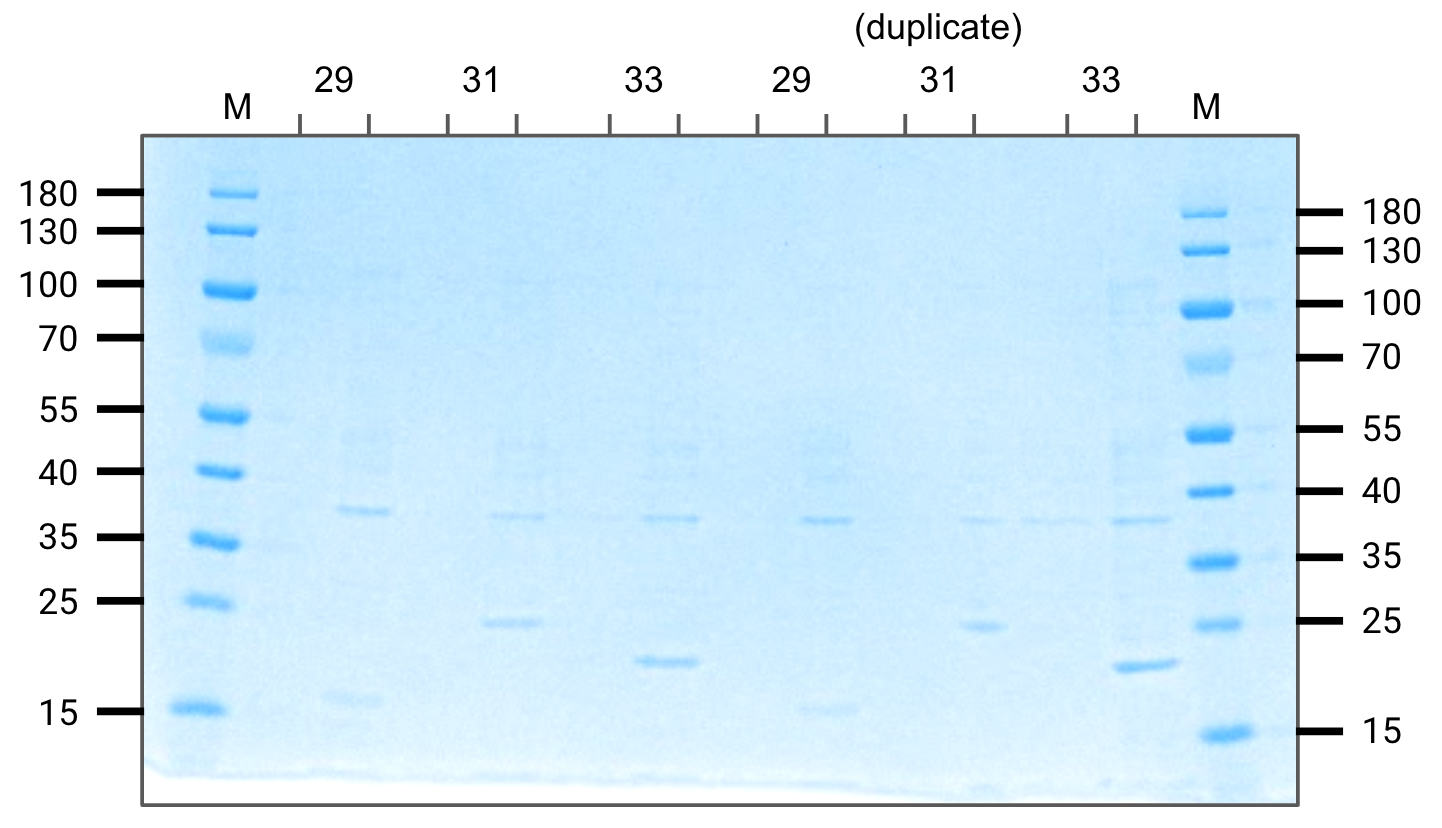
Expression gel (SDS-PAGE)
For each, left=uninduced, right=induced
Week 10
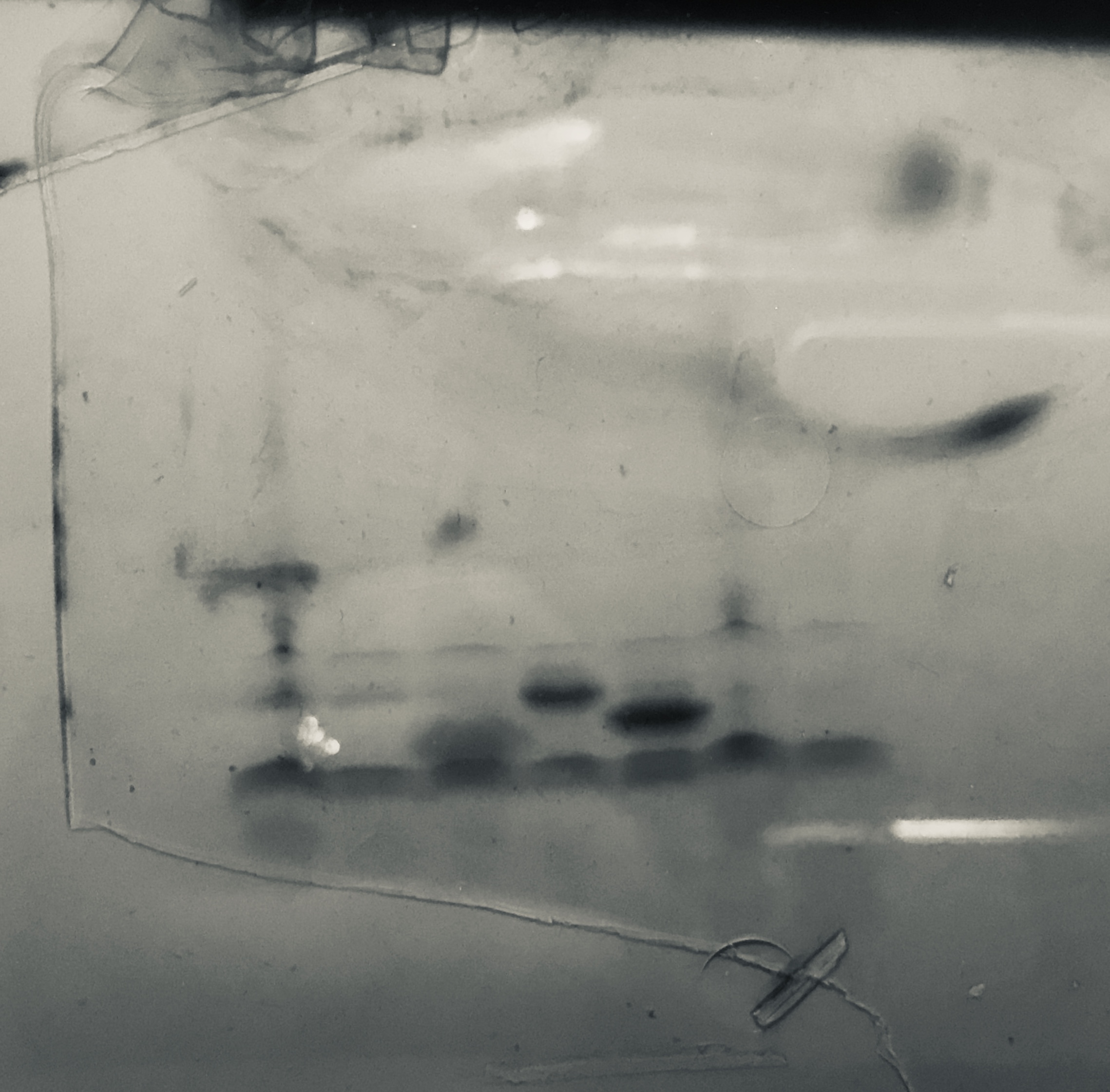
We tried purification of pVS31 and pVS33. Unfortunately most of our protein was retained in the pellet indicating insolubility.
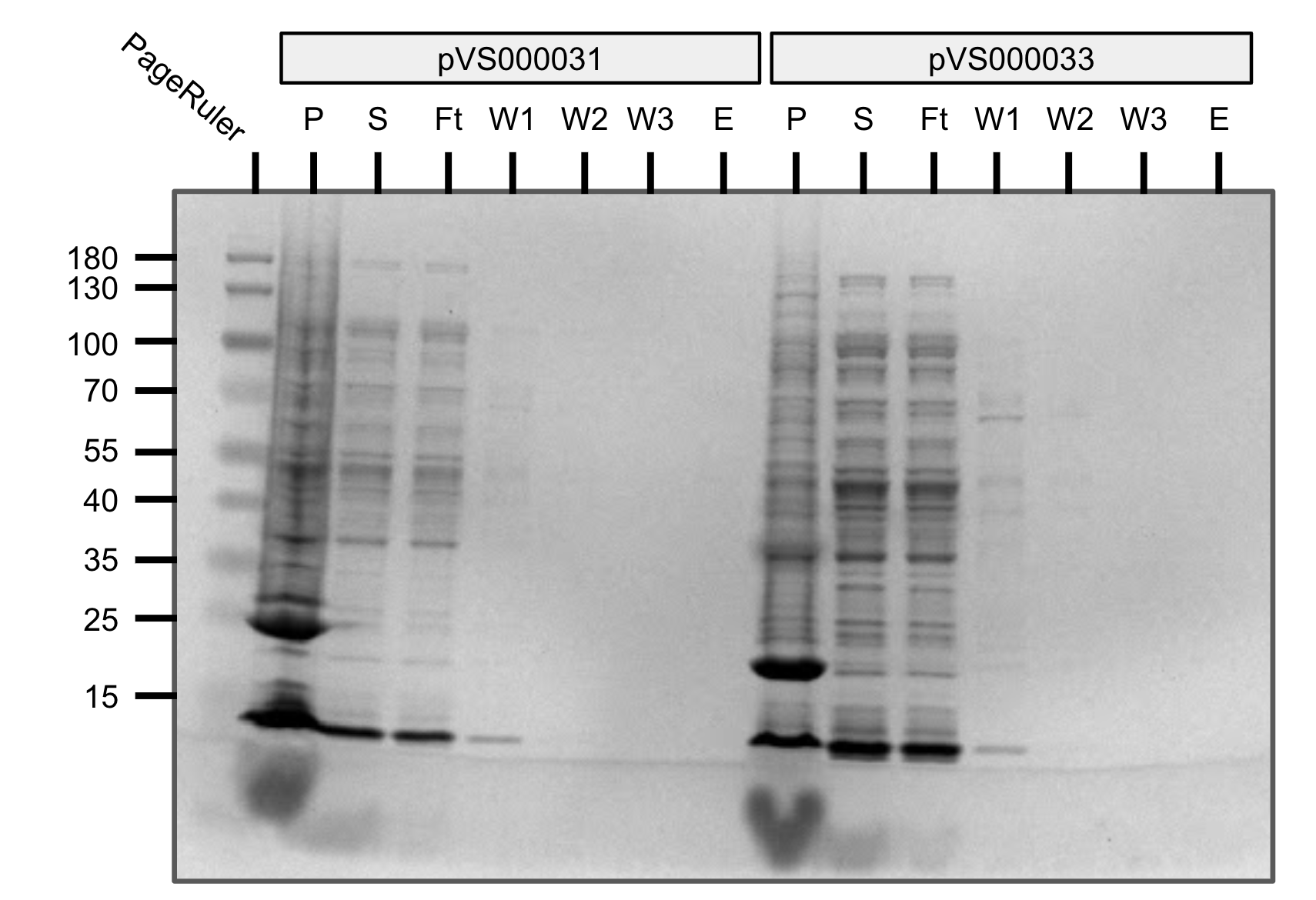
Purification gel (SDS-PAGE)
P=pellet, S=supernatant, Ft=flow-through, W=wash, E=elution
Attempted purification of pVS31 and pVS33 with an Emulsiflex. Most of the protein was retained in the pellet indicating solubility (as above).

Purification gel (SDS-PAGE)
Week 11
Since our previous expression tests were still relatively uncertain, we tried incubating to a higher OD (0.5-0.6) and expressing for 5 hours at room temperature.
We expressed set 1 constructs pVS(28,29,30,31,33,34,43) as well as our newly transformed set 2 constructs pVS(20,22,23,24,25,26).
Everything expressed at the expected size except for pVS28 (which did not express). After looking back at the sequencing results, we realized that our Gibson assembly introduced a deletion in the reading frame leading to a frame shift mutation.
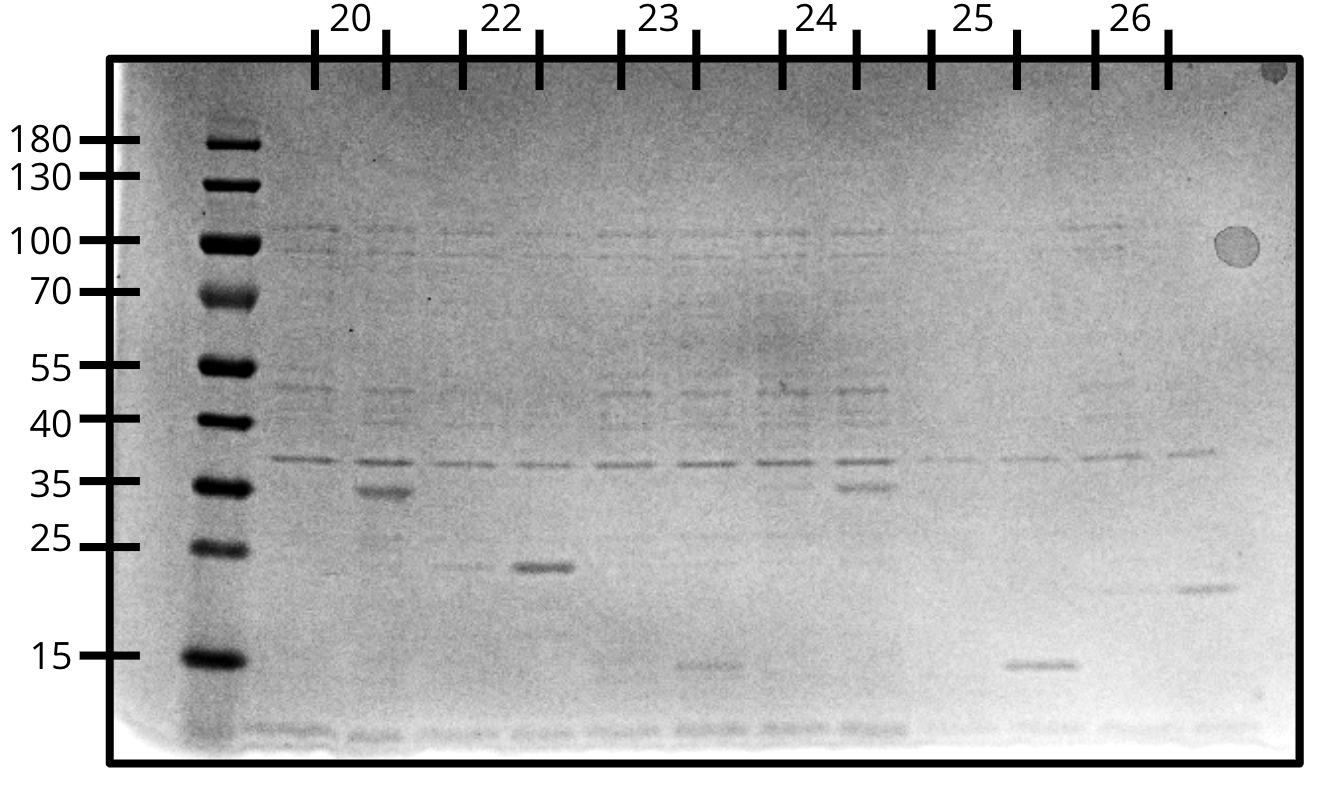
Expression gel (SDS-PAGE)
- pVS20: 32.02 kDa
- pVS22: 21.44 kDa
- pVS23: 14.90 kDa
- pVS24: 32.02 kDa
- pVS25: 13.62 kDa
- pVS26: 20.75 kDa

Expression gel (SDS-PAGE)
- pVS28: 19.86 kDa
- pVS29: 18.80 kDa
- pVS30: 18.69 kDa
- pVS31: 24.86 kDa
- pVS33: 19.91 kDa
- pVS34: 21.85 kDa
- pVS43: 21.44 kDa
Since several of our constructs appeared to be relatively insoluble, we decided to try cloning our inserts into pTEV6 which expresses maltose binding protein as an N-terminal fusion tag.
To extract our insert, we ordered insertion primers and ran PCR followed by gel electrophoresis. Then we cut out the corresponding bands and extracted the DNA.
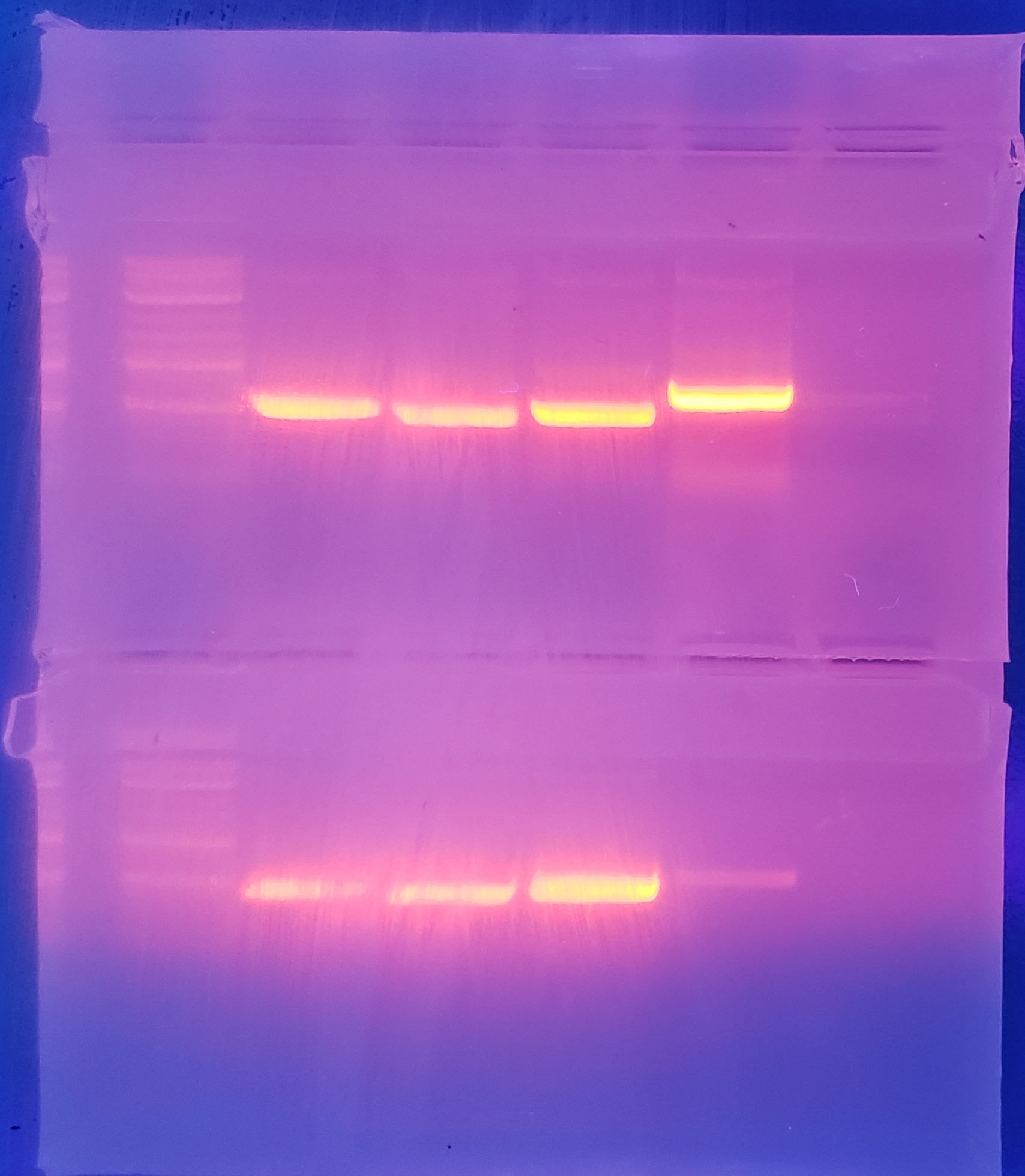
Insertion bands
Top: pVS(28,29,30,31)
Bottom: pVS(33,34,43)
We expressed 250mL cultures of pVS(29,30,34,43) and pelleted to prepare for further purification.
We attempted Ni-NTA purification of three constructs and finally figured out the protocol.
All three constructs expressed and showed up in the elution with minimal contaminants.
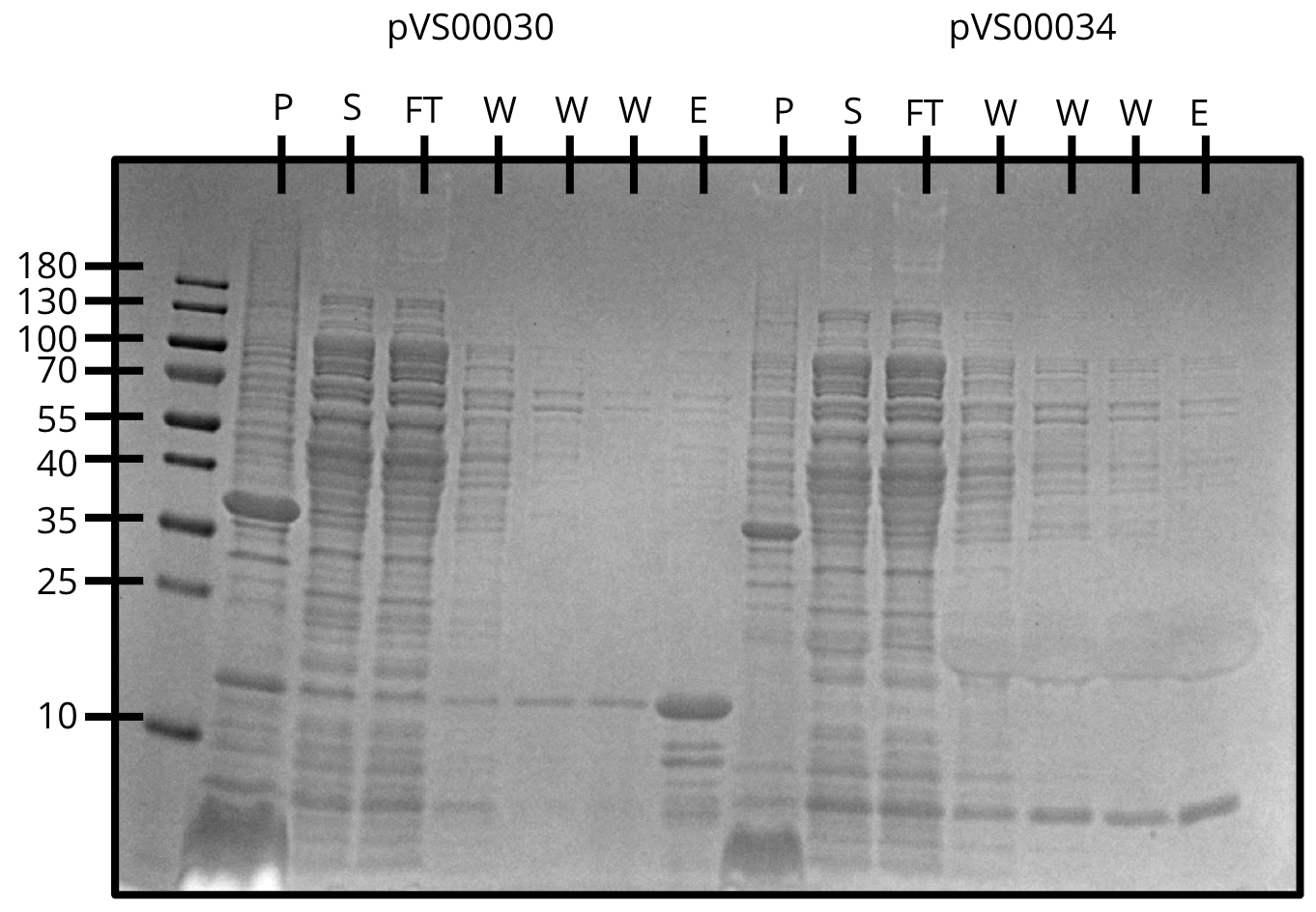
Ni-NTA purification of pVS30 and pVS34
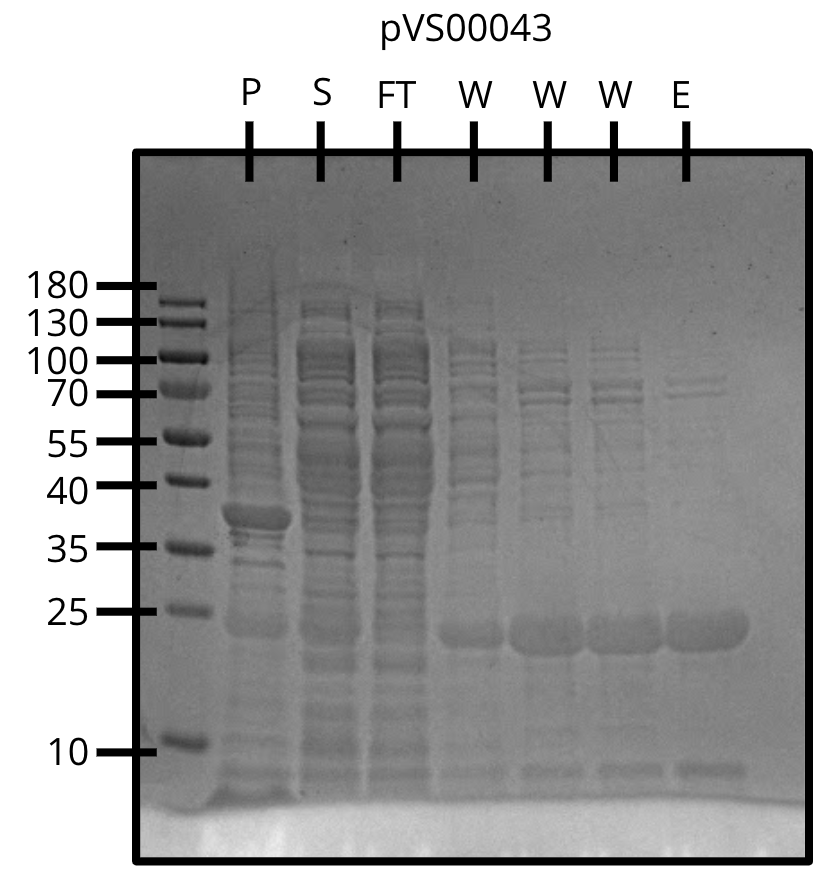
Ni-NTA purification of pVS43
Week 12
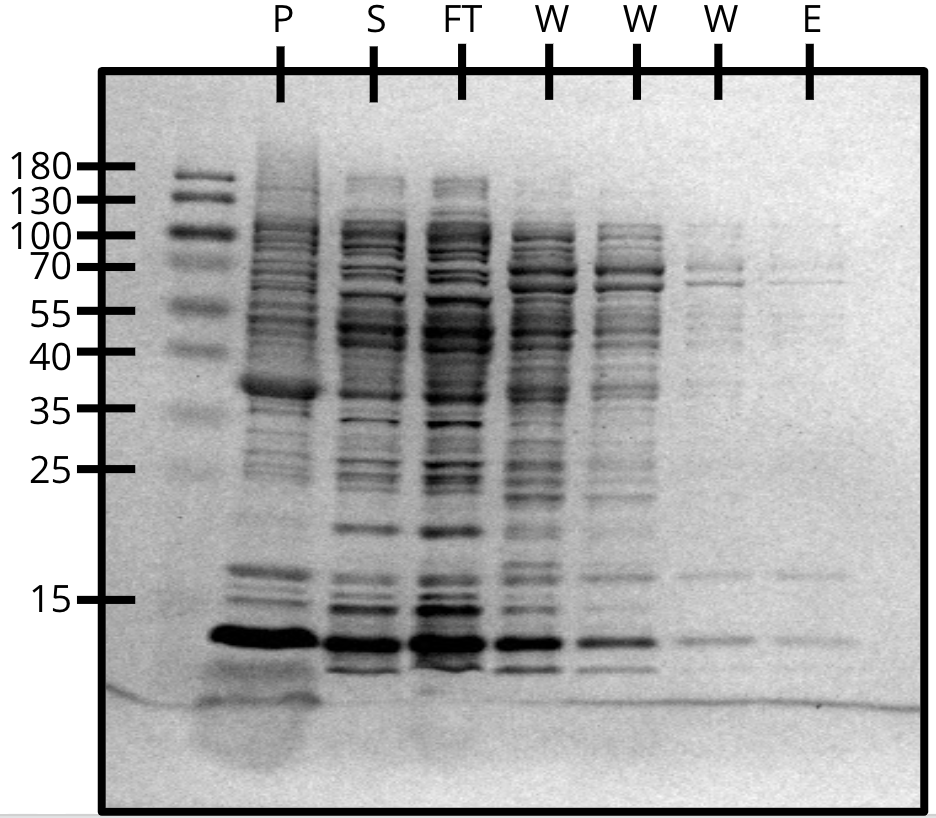
Ni-NTA purification of pVS29

Ni-NTA purification of pVS22(left) and pVS23(right)
pVS22 seems to be relatively insoluble.
Week 13
We were able to purify all the cosntructs with varying levels of purity.
U=uninduced, I=induced, P=pellet, S=supernatant, Ft=flow-through, W=wash, E=elution
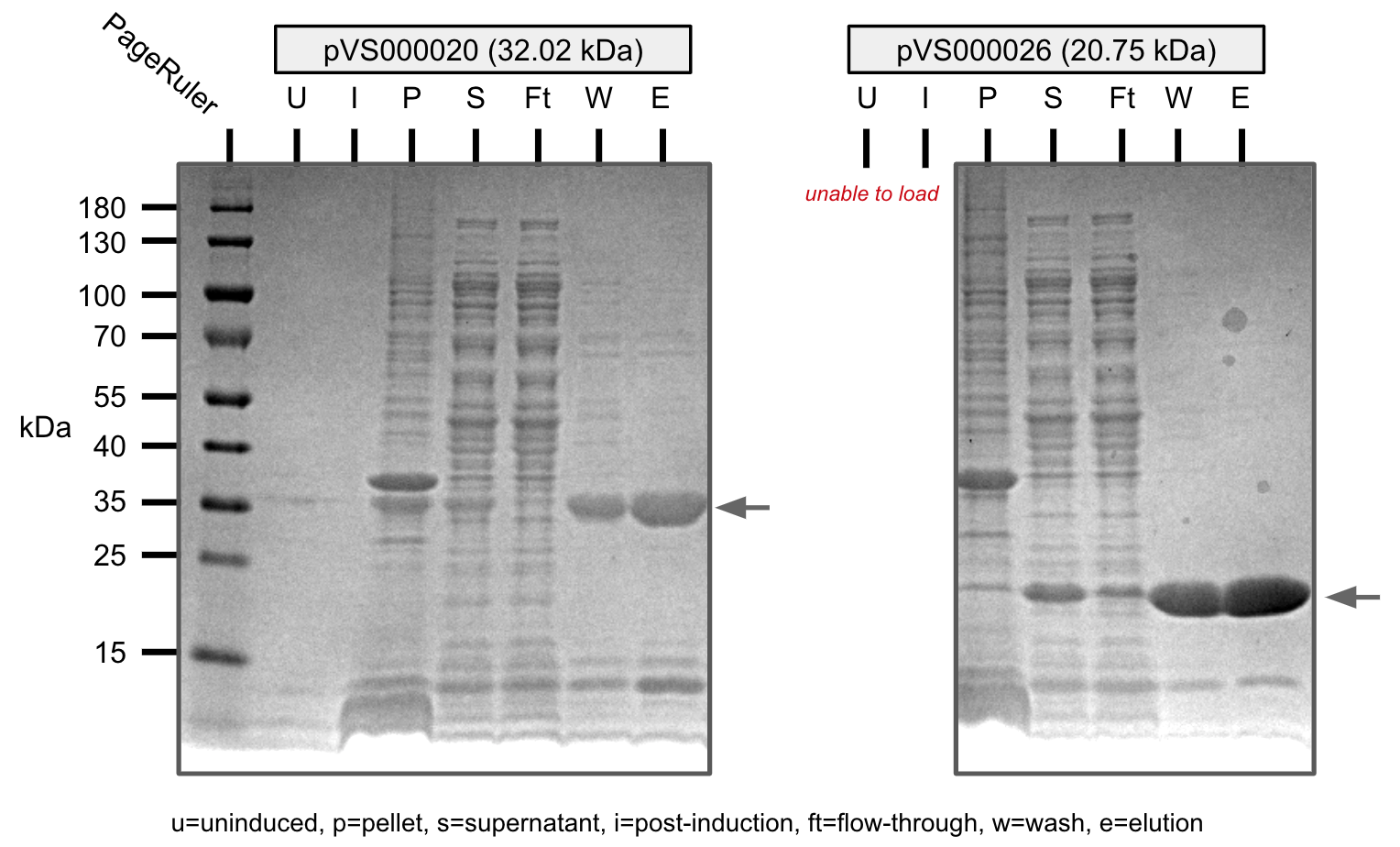
Ni-NTA purification of pVS20 and pVS26 (SDS-PAGE)
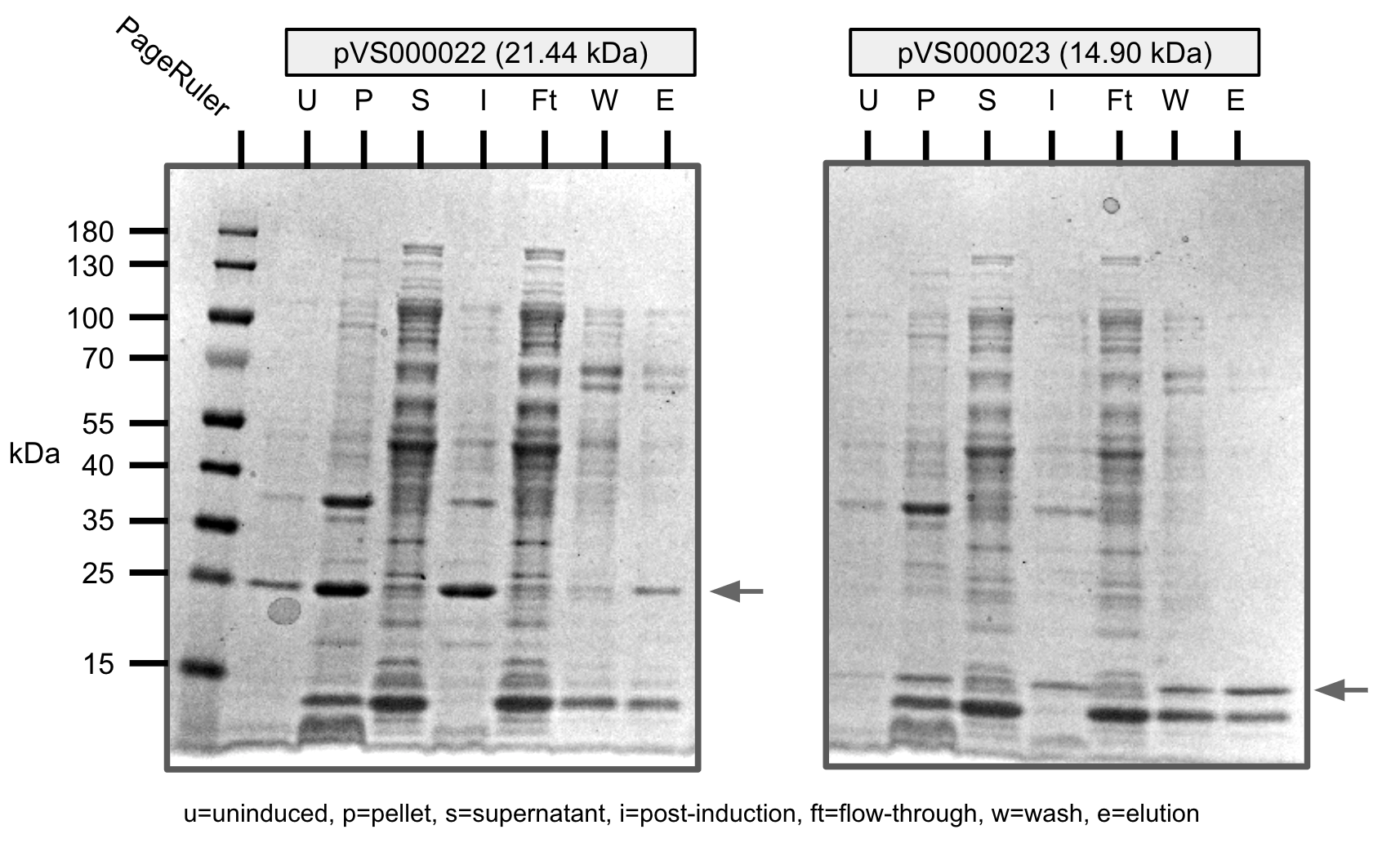
Ni-NTA purification of pVS22 and pVS23 (SDS-PAGE)
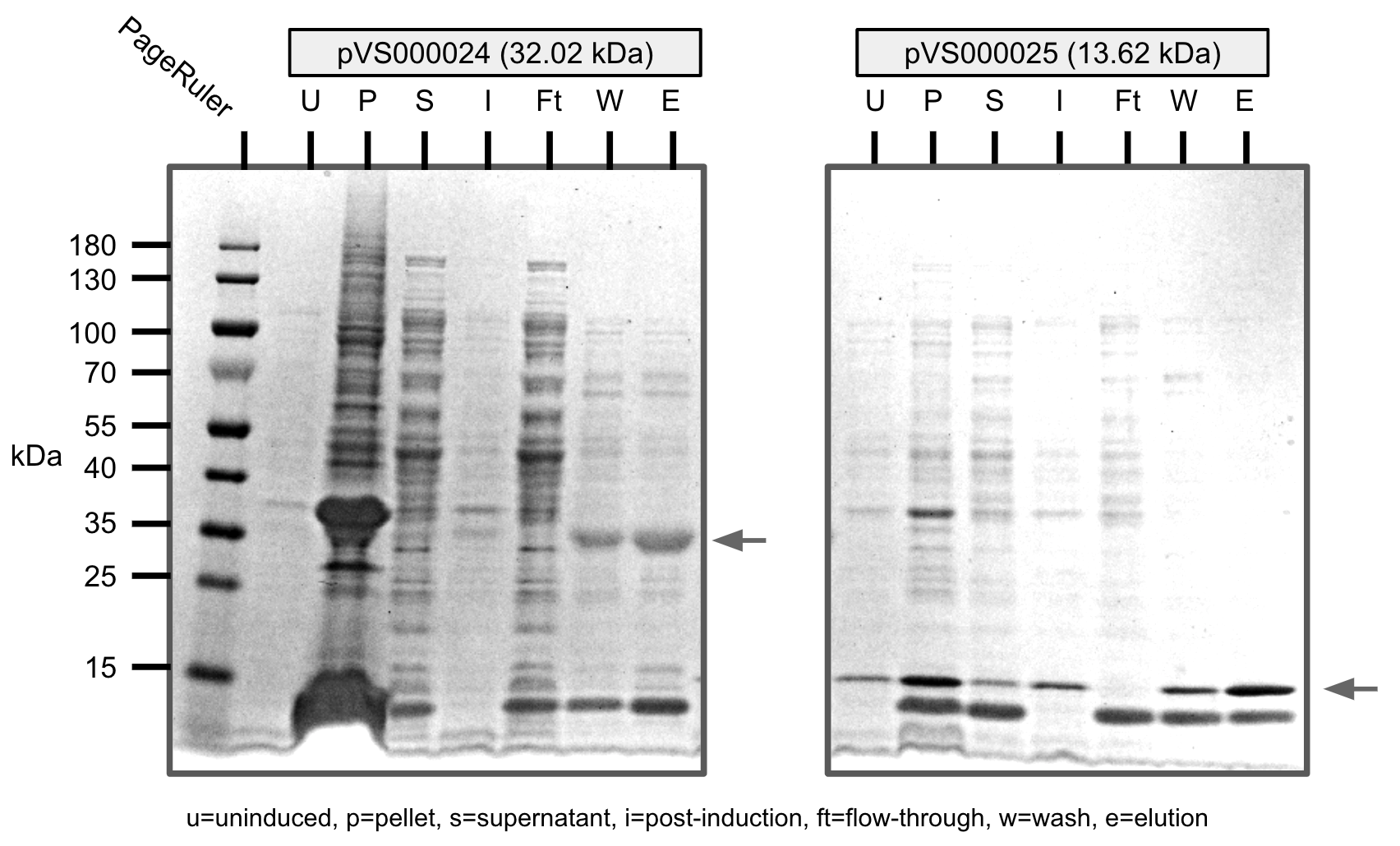
Ni-NTA purification of pVS24 and pVS25 (SDS-PAGE)
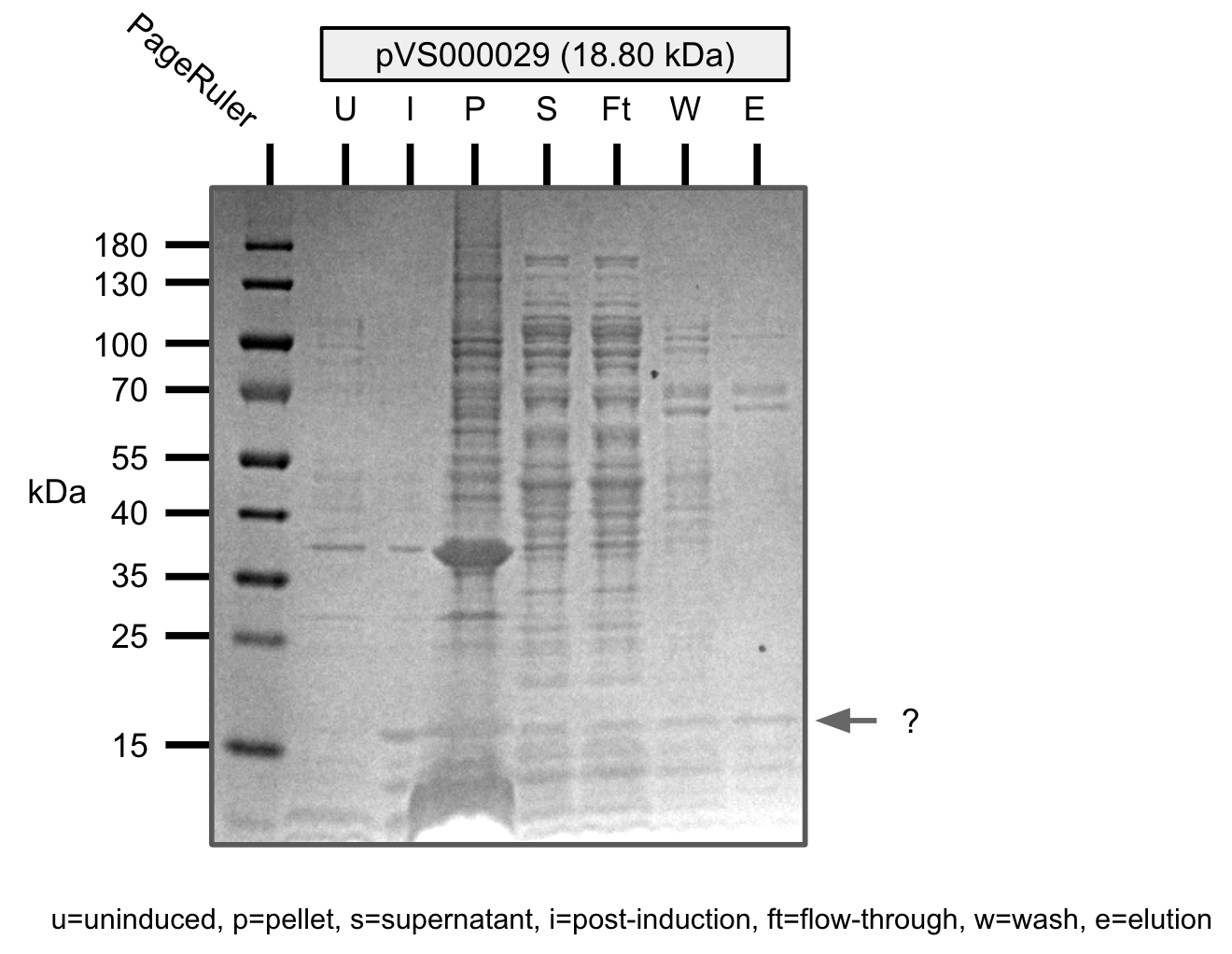
Ni-NTA purification of pVS29 (SDS-PAGE)
After observing no colonies growing on AMP+ LB plates, we decided to retry colony PCR to verify the insertion.
Unfortunately, we didn't see a single positive hit from these colonies, indicating that cloning did not work.
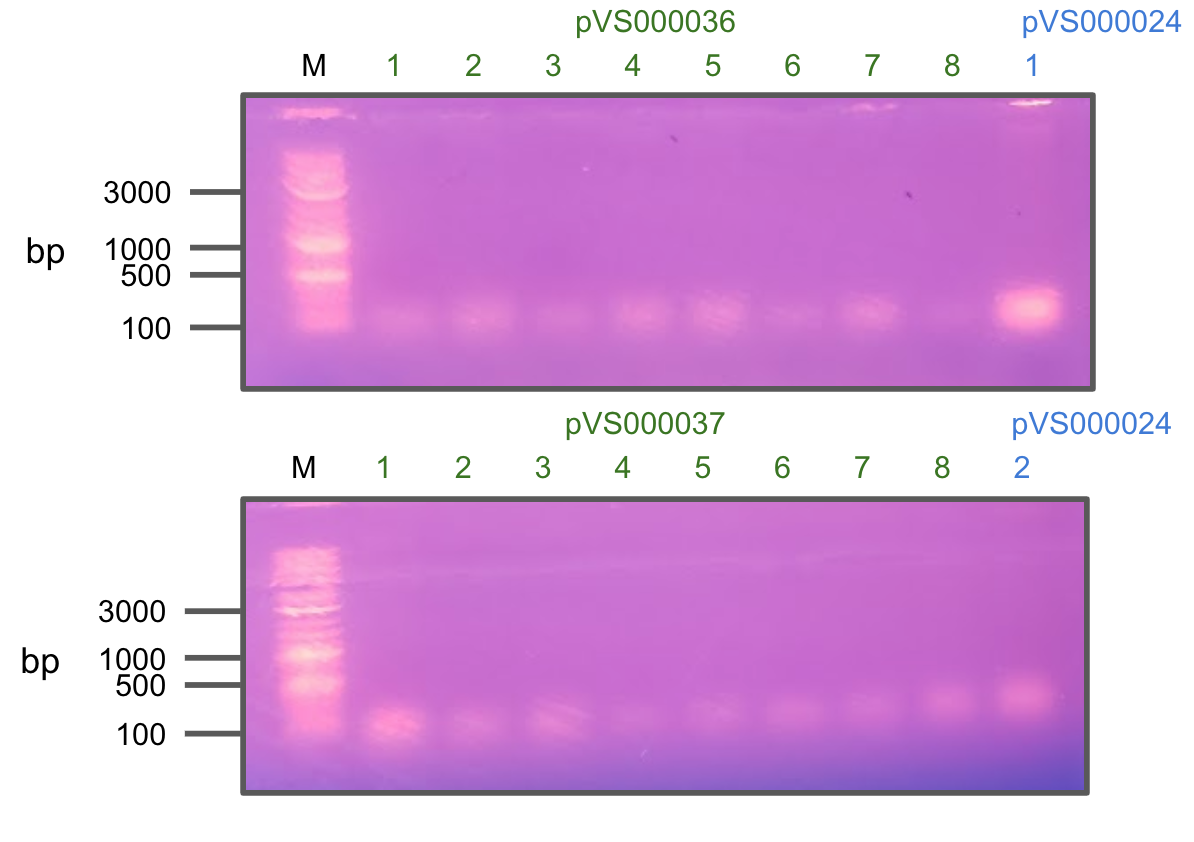
Colony PCR of pVS36 and pVS37 (+pVS21)
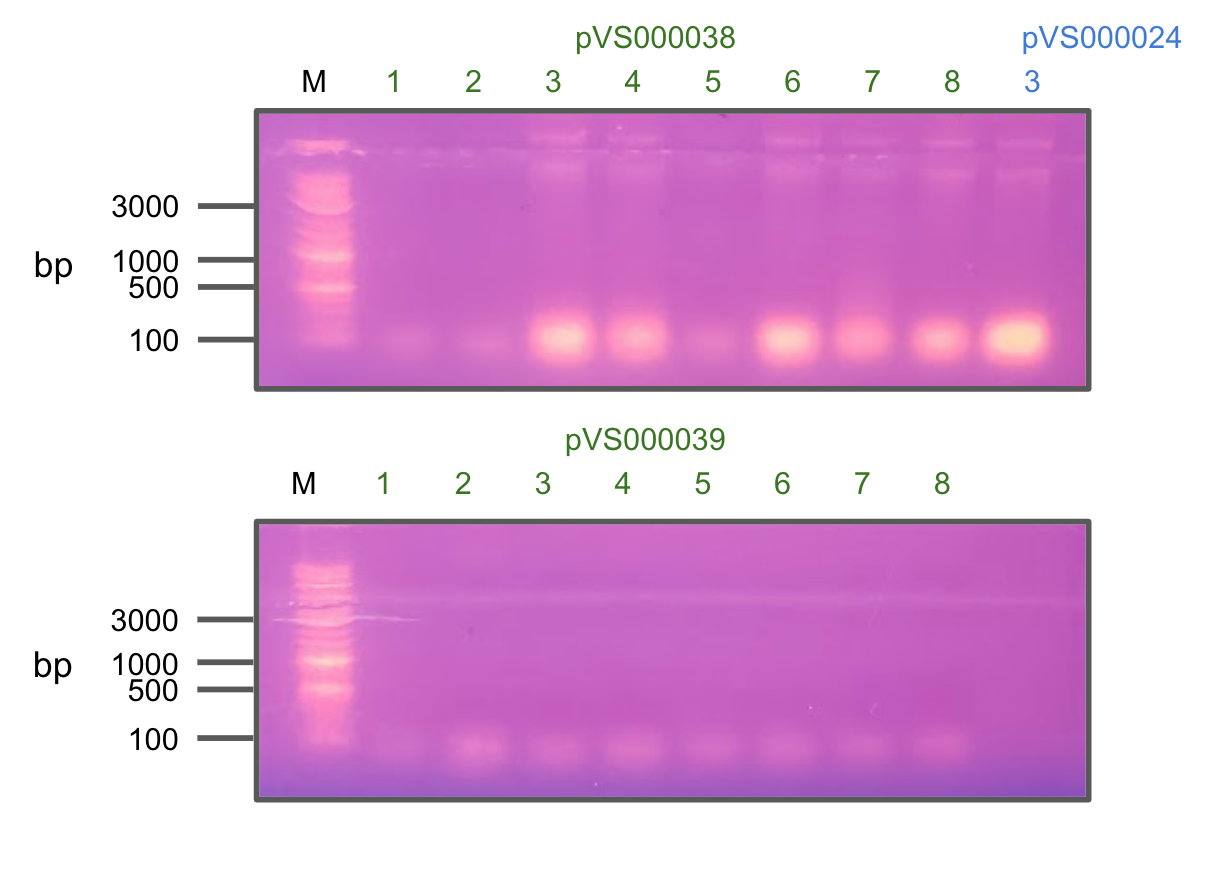
Colony PCR of pVS38 and pVS39 (+pVS21)
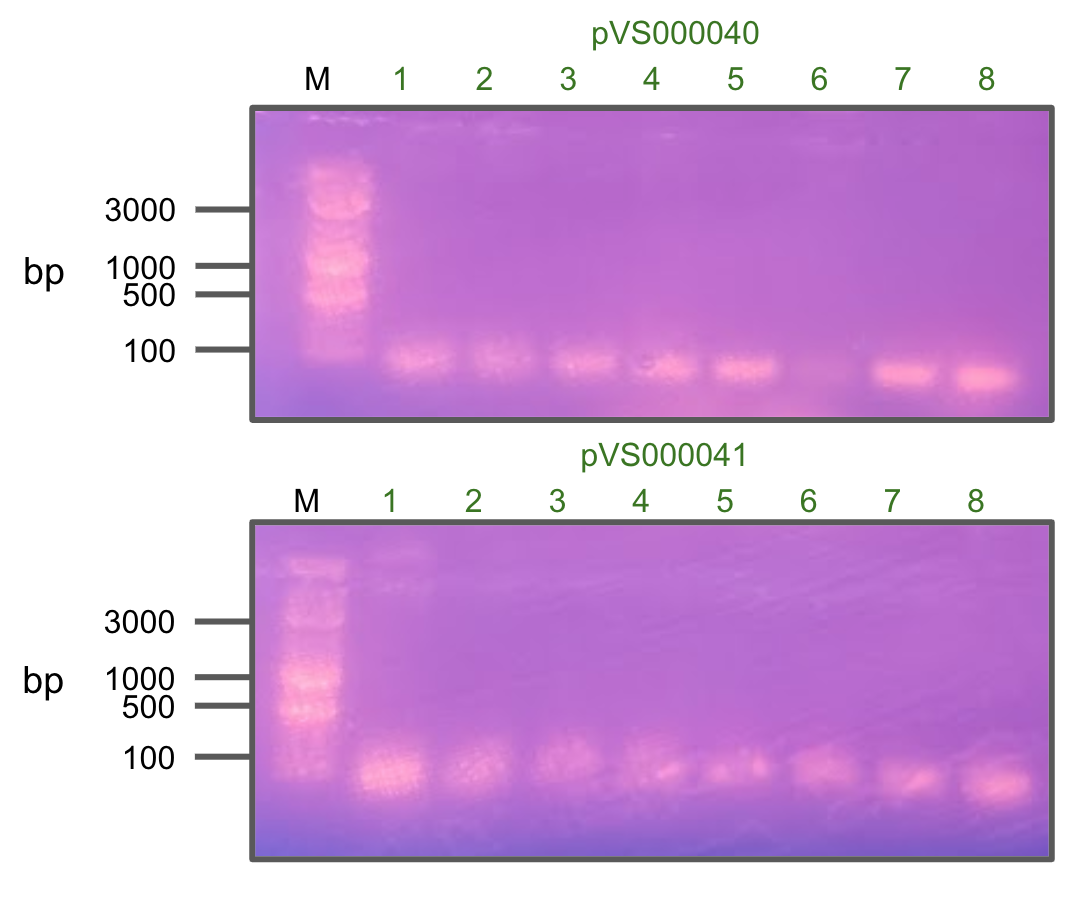
Colony PCR of pVS40 and pVS41
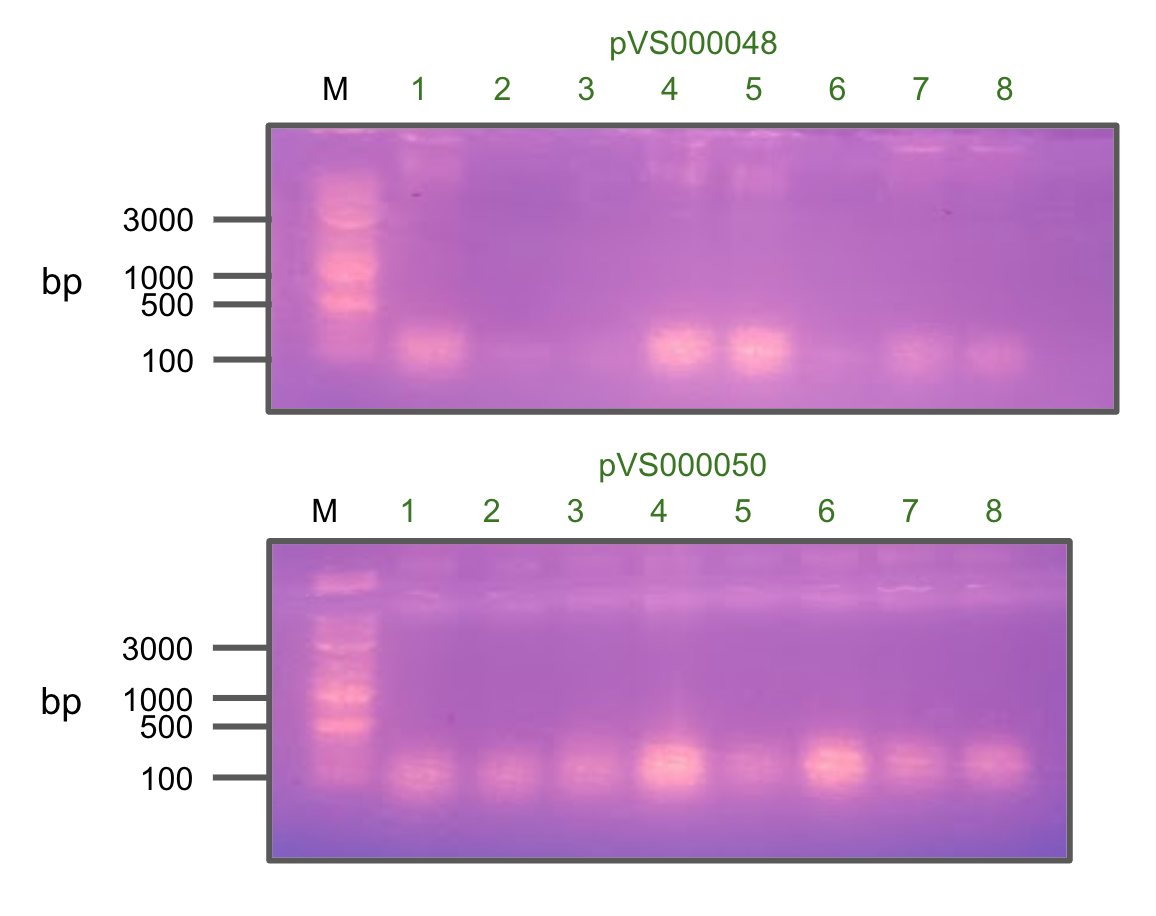
Colony PCR of pVS48 and pVS50
We attempted to mix whole-cell lysates containing each of our intein halves to try to observe intein splicing.
However, background cellular proteins made it very difficult to identify any bands that were formed or depleted.
In the future, we could try using a His-tag stain to identify our synthetic protein constructs (and reactants).
Miniprepped two other colonies of pVS28 and sent in for sequencing.
Week 14, 15, 16
Everyone is on break
Week 17
Attempted purification of pVS33 with BugBuster.
Protein did not show up in the elution.
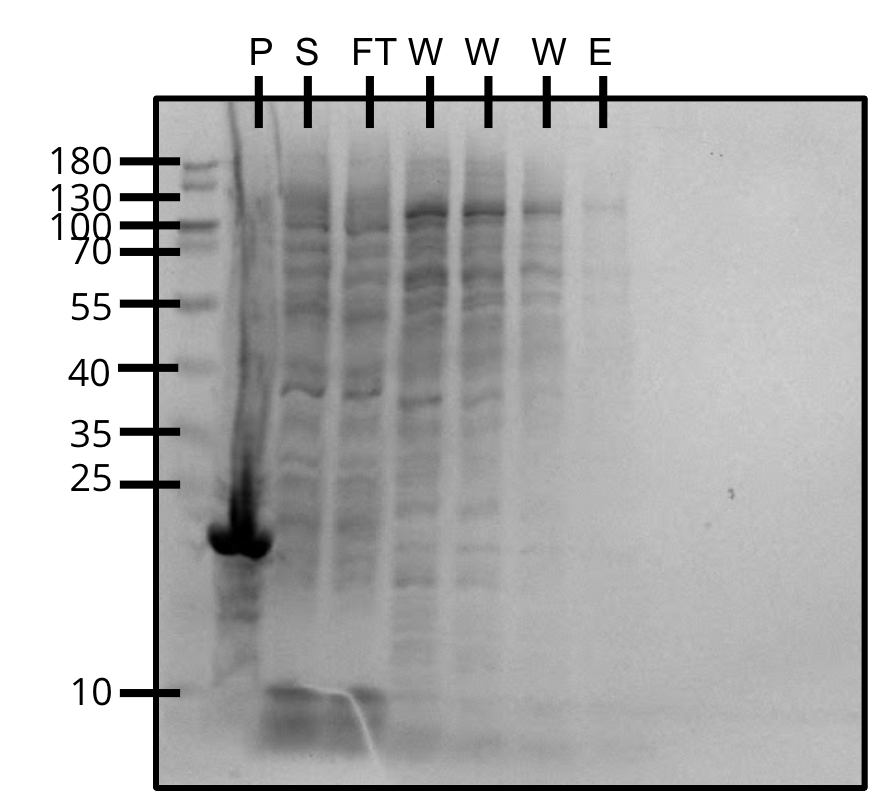
Purification of pVS33 (SDS-PAGE)
Week 18
Attempted Gibson cloning of our set 1 constructs in pTEV6 and verified insertions via colony PCR.
Transformed several constructs from CSL into BL21 for expression.
Gibson assemblies were transformed into DH5α comp cells.
Prepared more BL21 comp cells for future transformations.
Two constructs encoding sspDnaB split-inteins were transformed into E. coli BL21 for expression and colony PCR was used to verify inserts.
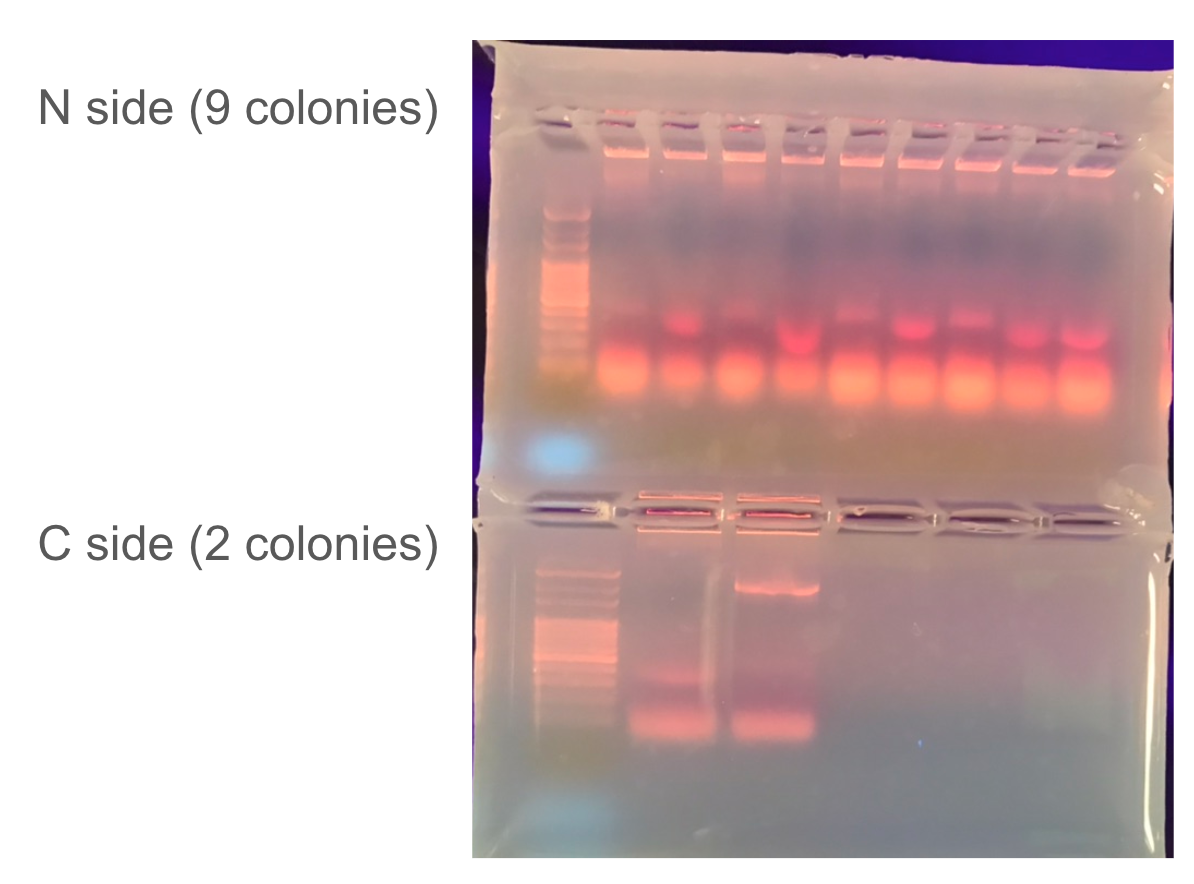
Colony PCR
Attempted purification of pVS29 and pVS30 with BugBuster and sonic dismembrator as a control.
Gel overstained, it is difficult to tell which method worked better.
Week 19
Plates containing our existing constructs were transported to our secondary workspace in Dr. Childer's lab in order to make use of their equipment. These constructs were miniprepped and retransformed into DH5α comp cells.
DH5α constructs from 9/16 were miniprepped and transformed into BL21 for expression.
Additonally a second set of transported constructs was miniprepped and transformed into DH5α.
The second set of transported constructs was minipreppe from DH5α colonies and transformed into BL21 comp cells.
We received gBlocks encoding our split-linker constructs from IDT and cloned them into a pTEV6 backbone. Gibson assemblies were transformed into E. coli DH5α cells and plated on LB media. We also plated several previous assemblies of our pTEV6 variants of set 1.
Made freezer stock of all the transported BL21 colonies for future expression.
Performed colony PCR on the newly transformed constructs in order to screen for insertions.
Used pTEV6 primers provided by a graduate student in Dr. Childer's lab.
Bold/underlined colonies indicate positive hits.
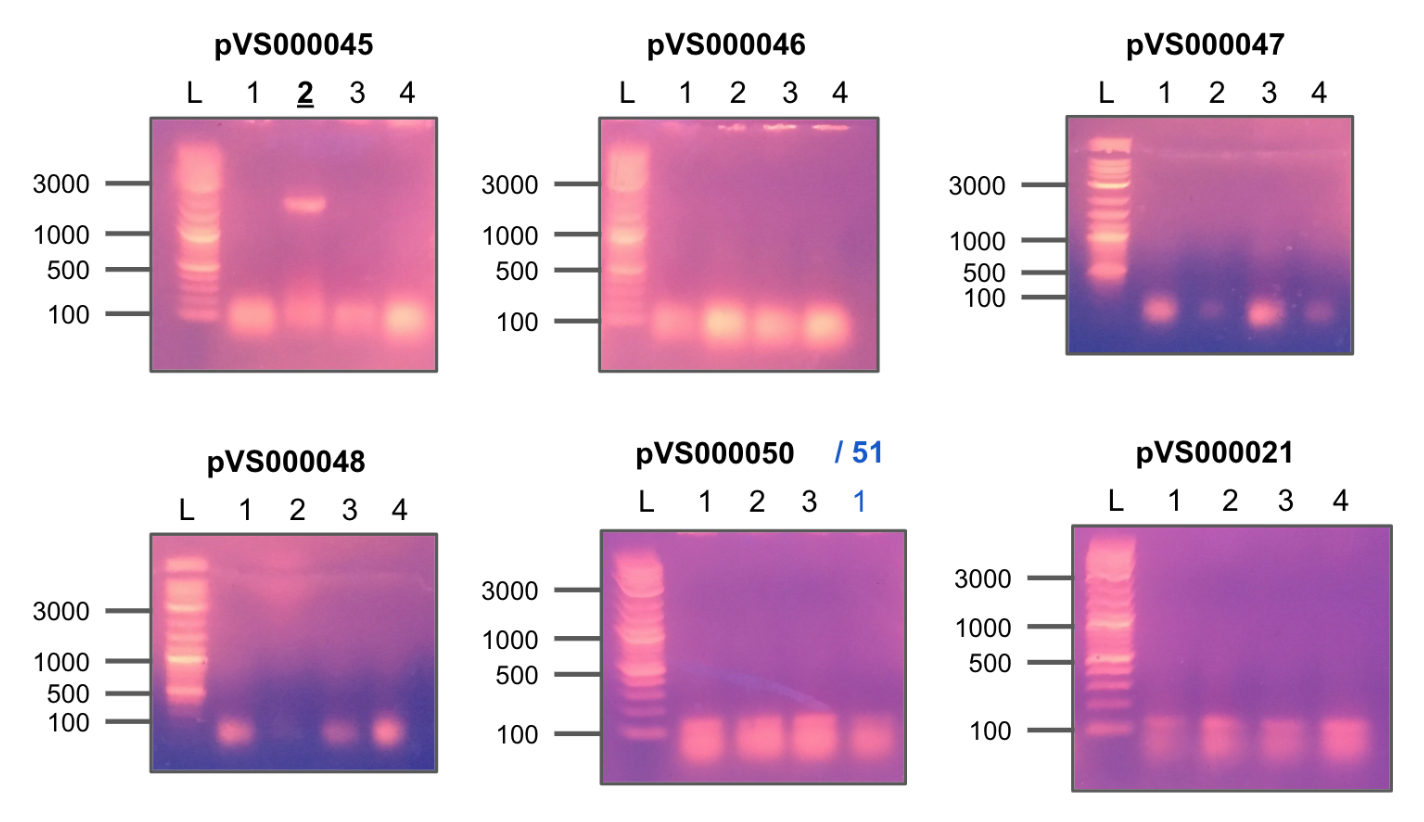
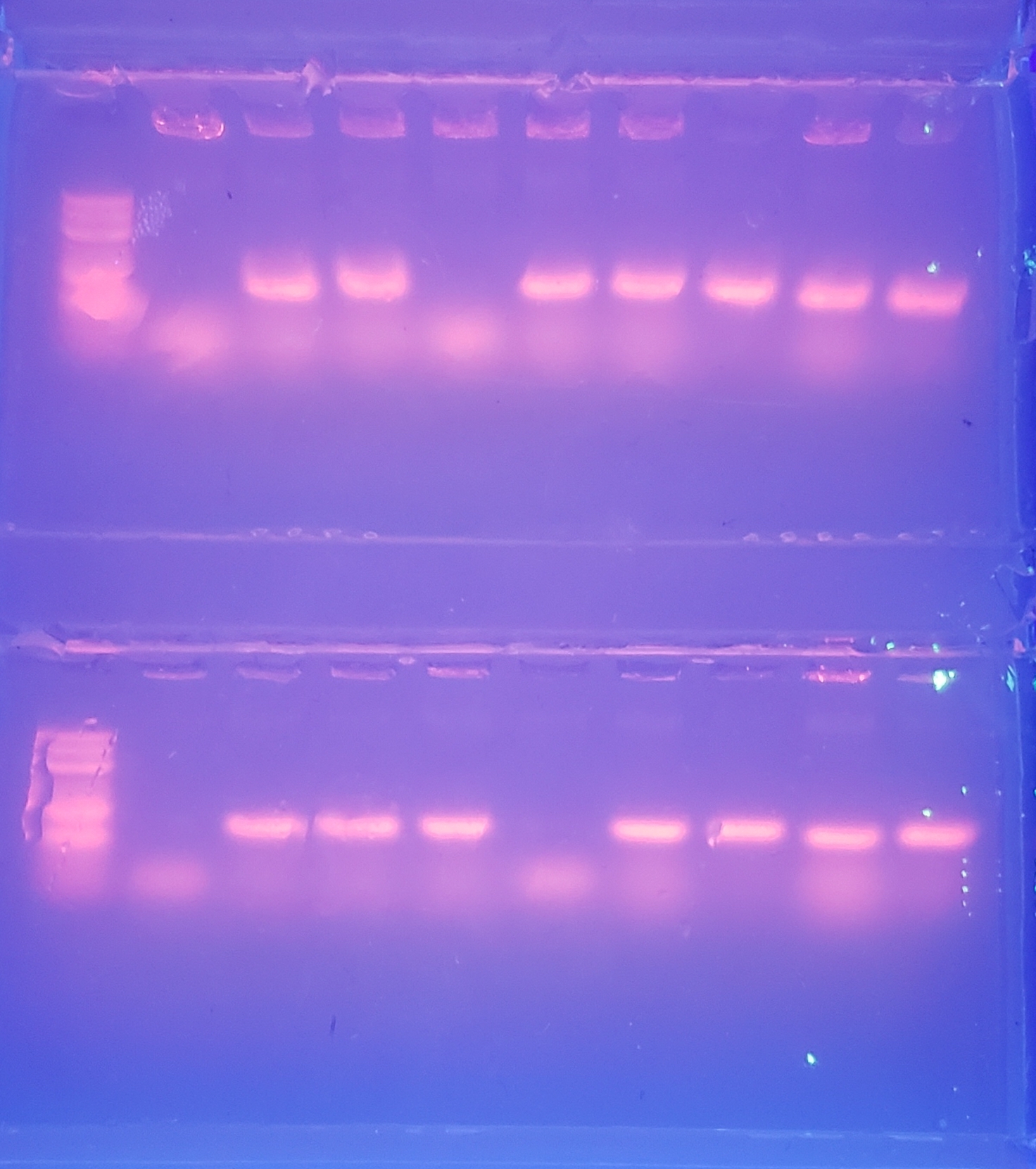
Colony PCR 1
Colony PCR 2
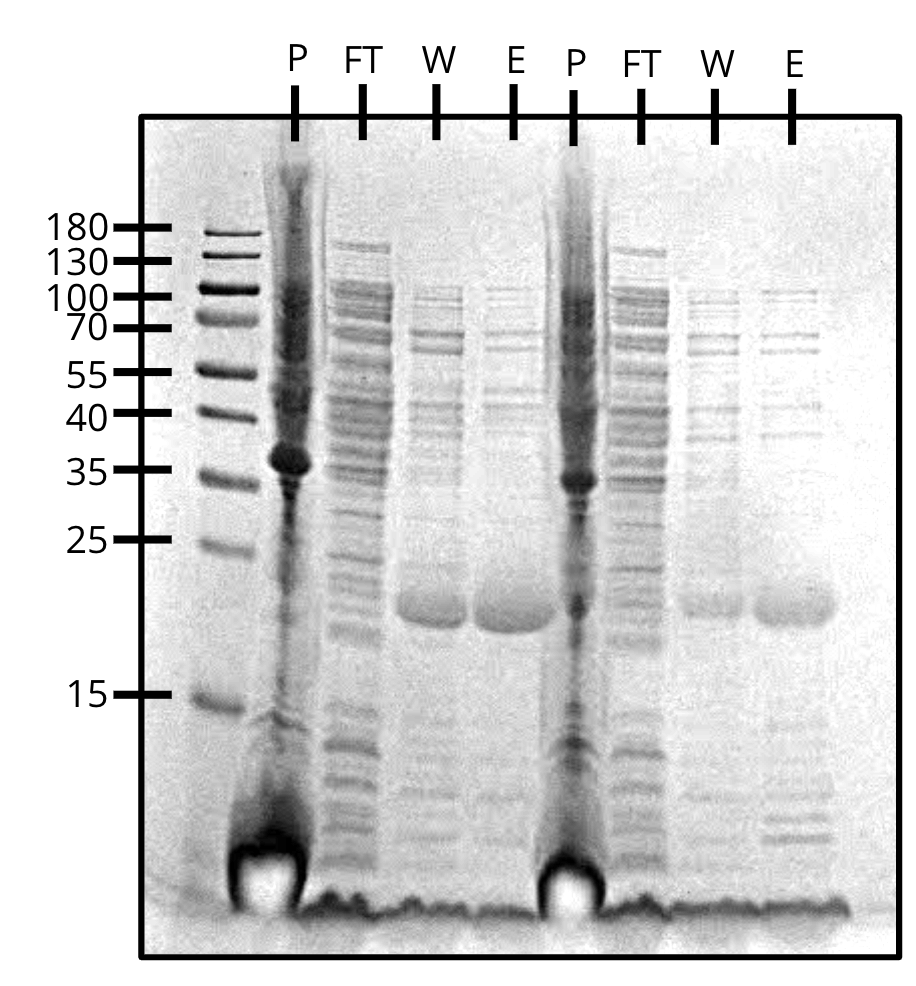
Purification of pVS34 (left) and pVS43 (right) (SDS-PAGE)
Week 20
Overnight colonies of the positive hits were miniprepped to prepare for sequencing.
Designed and ordered four new pTEV6 screening and sequencing primers to get better colony PCR results and prepare for Sanger sequencing.
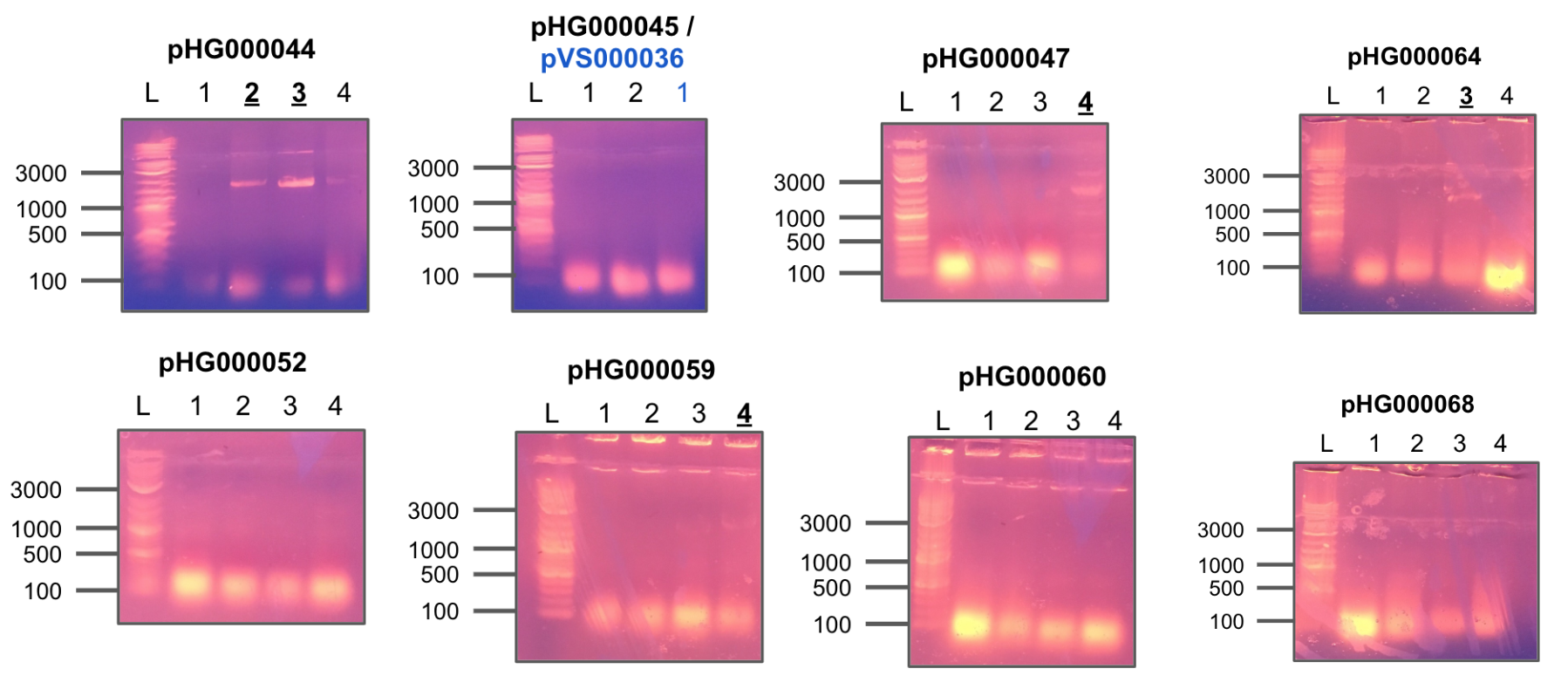
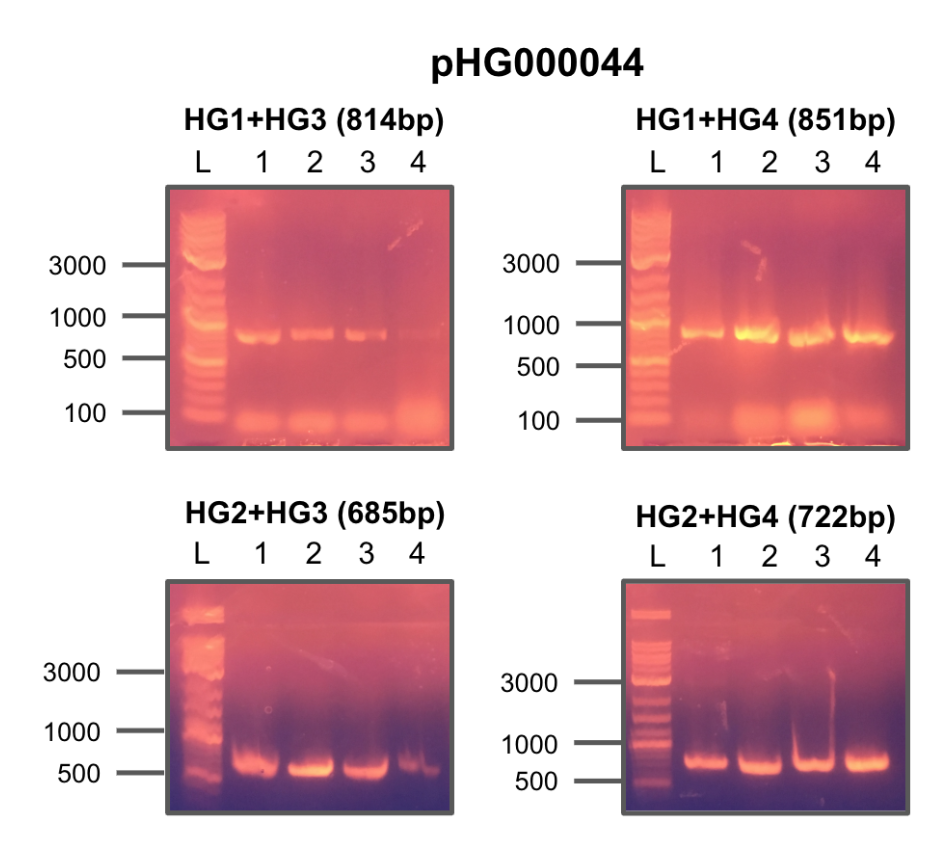
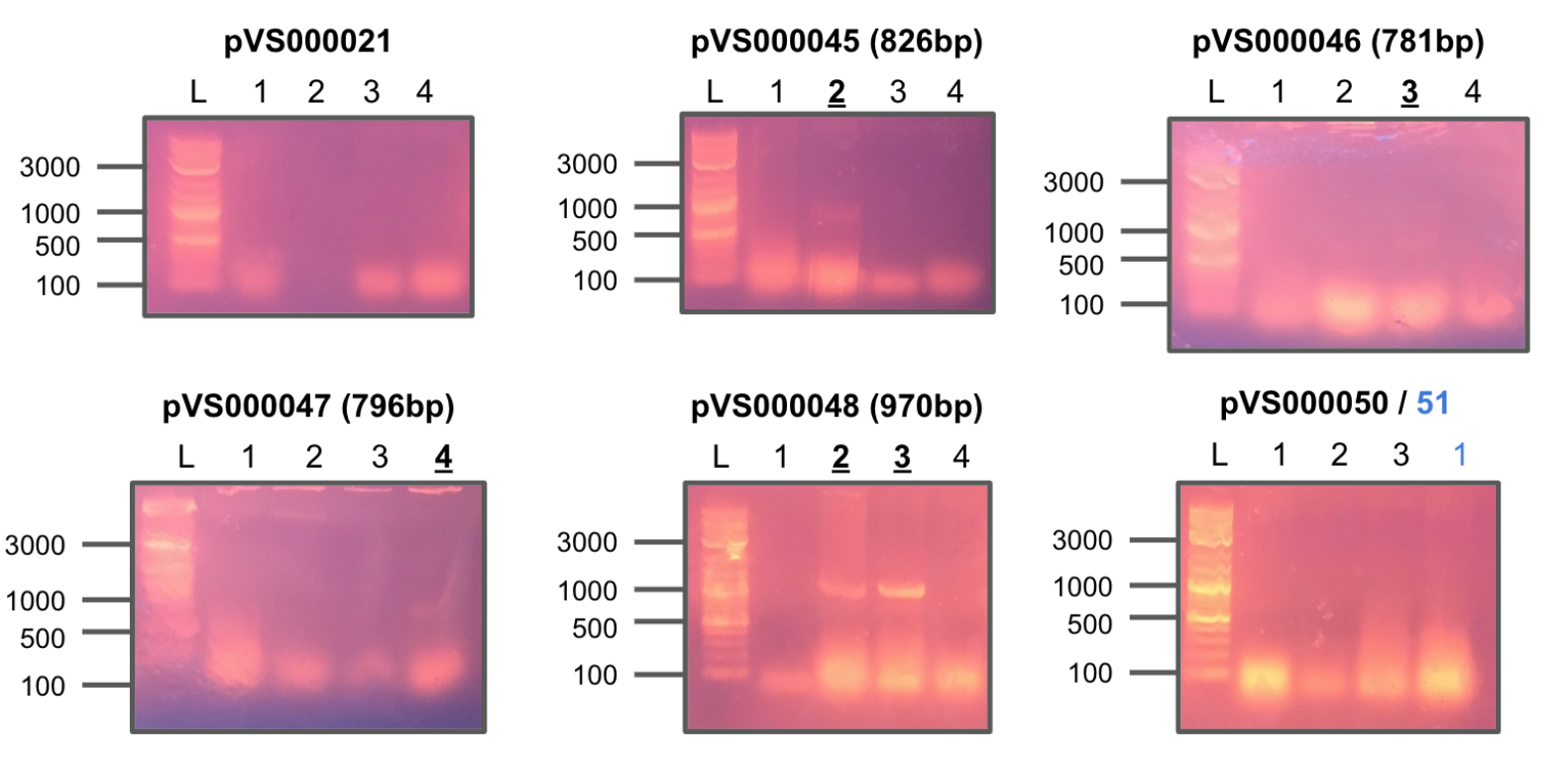
Received primers from IDT and tested each combination of F/R primers on four constructs of pHG44.
All colonies showed strong positive hits.
Colony PCR
Redid colony PCR of the previous constructs using the new pTEV6 primers.
Colony PCR 1
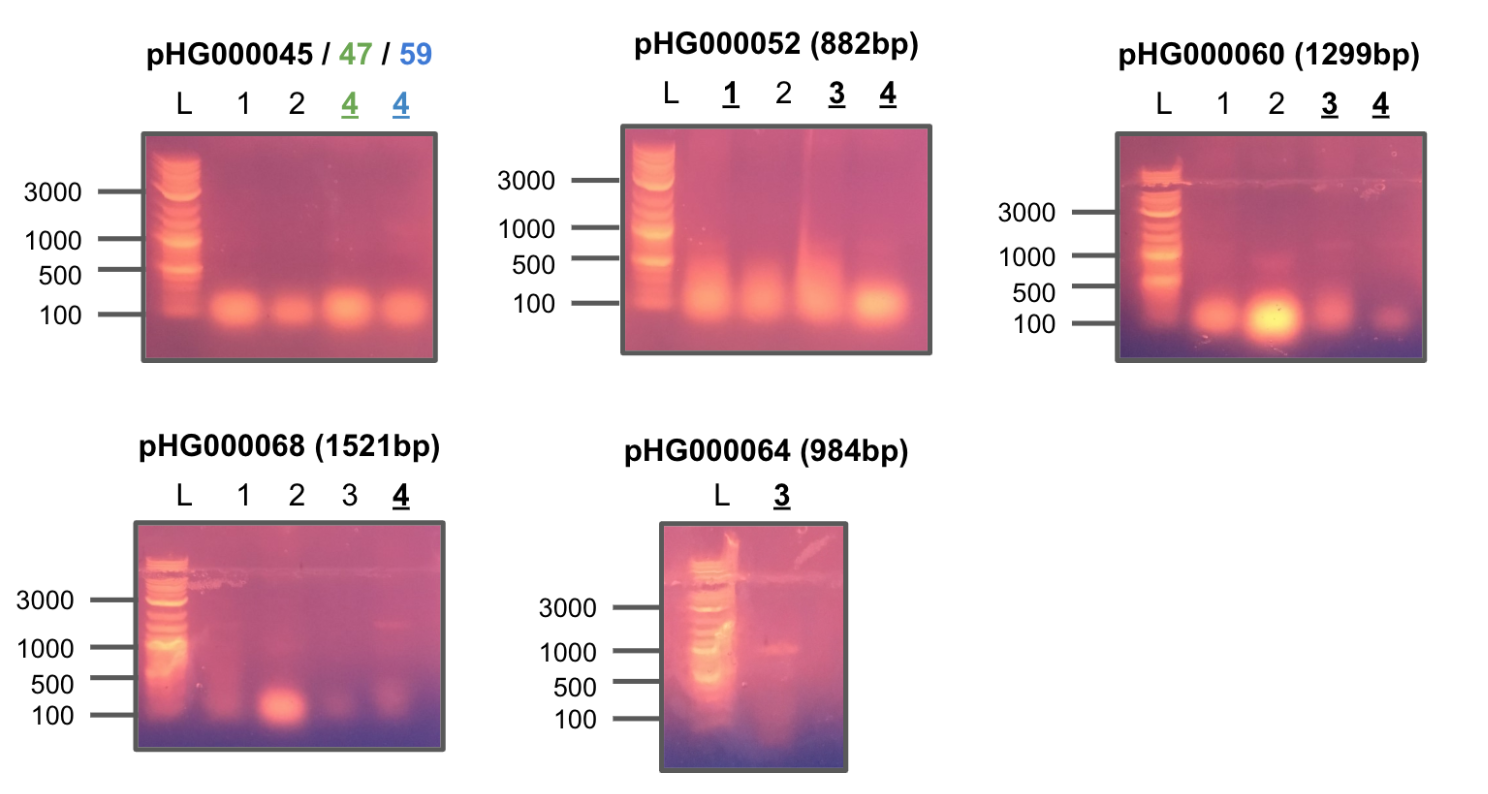
Colony PCR 2
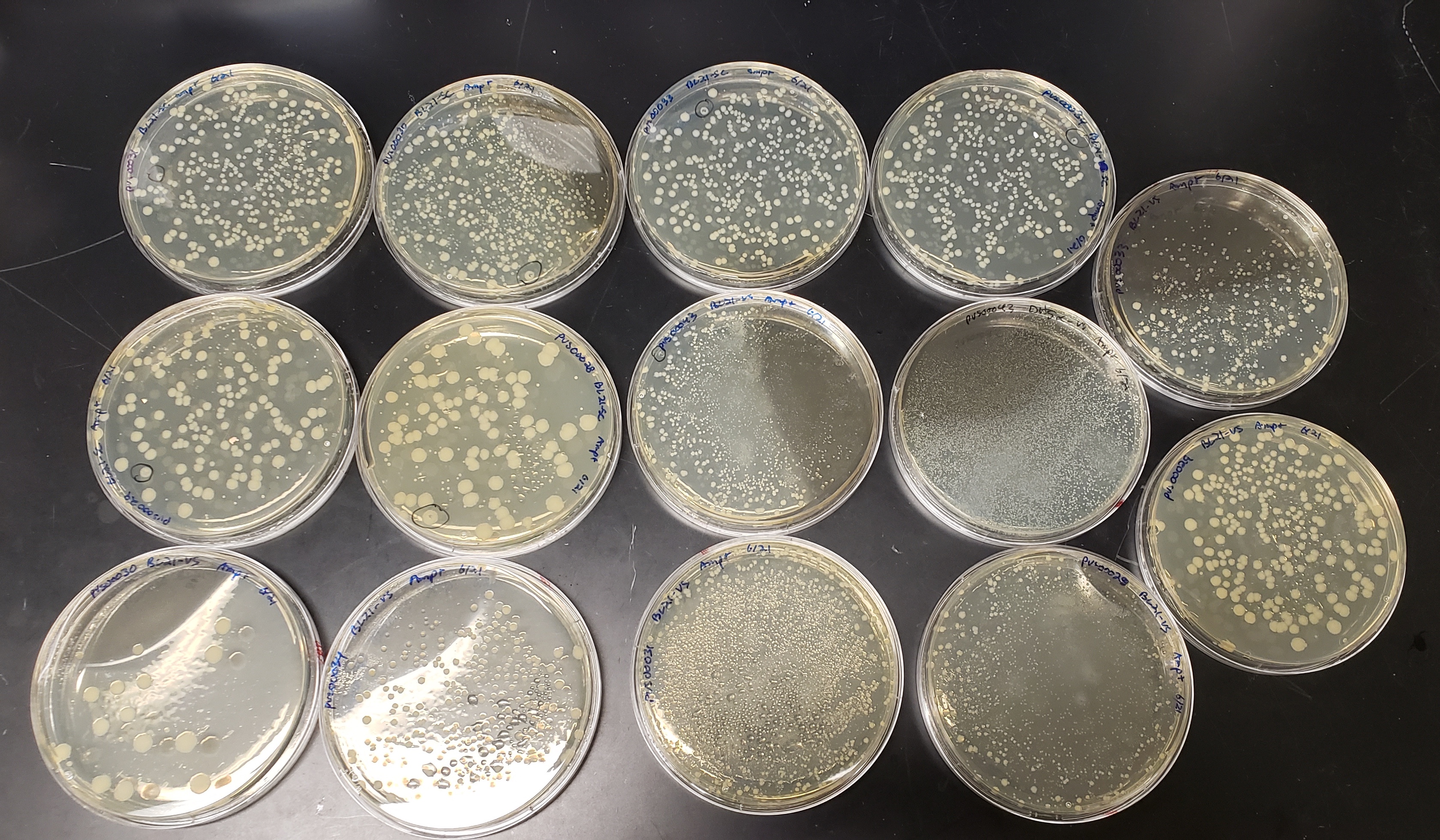
All the colonies with positive hits were miniprepped from overnight cultures and transformed into E. coli BL21 for expression.
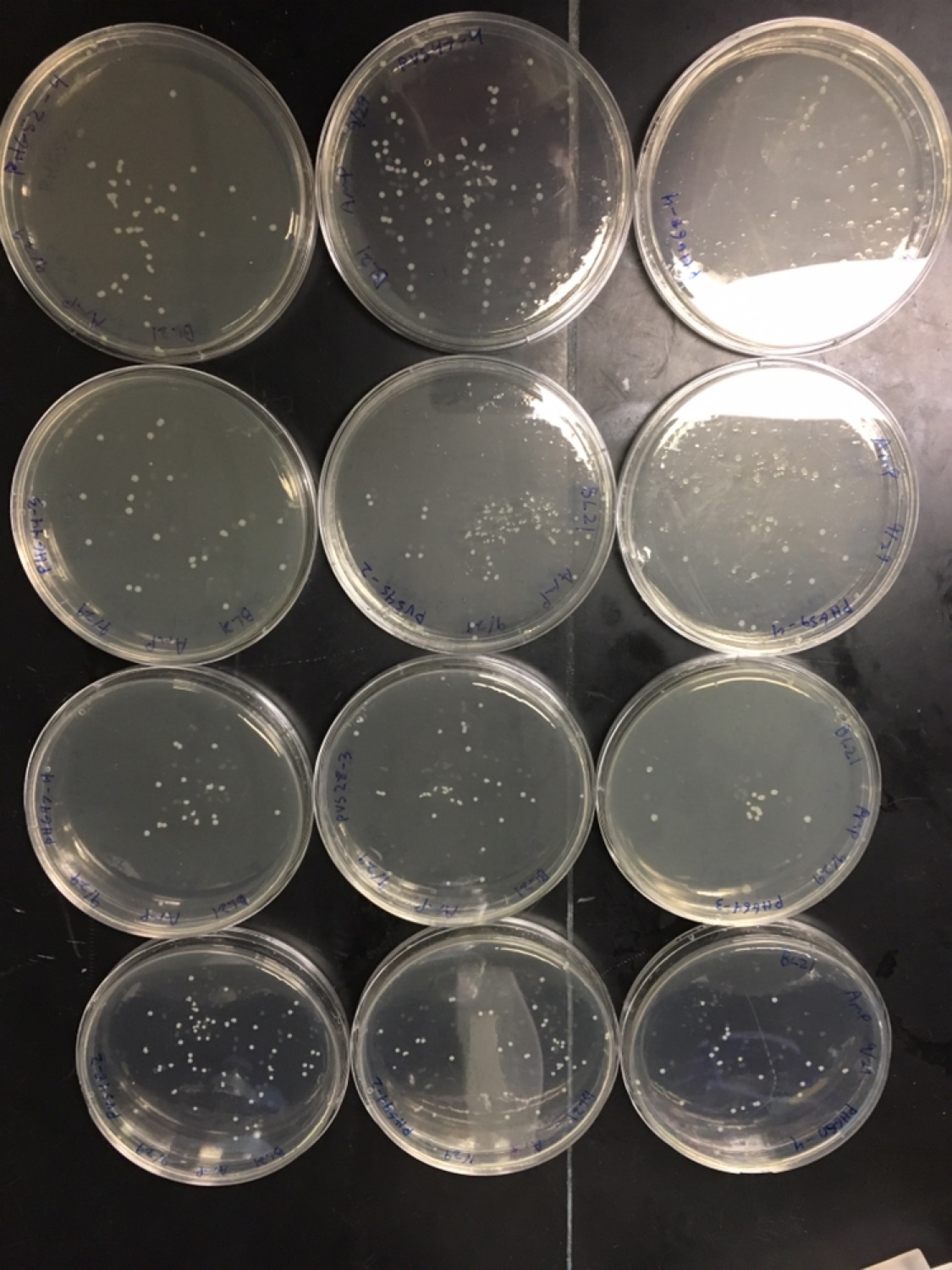
BL21 plates
Week 21
Large scale expression of pVS22 for lysate mixture tests.
Created freezer stock of newly transformed BL21 constructs from overnight cultures.
Grew 4mL cultures of the new constructs to OD600=0.5-0.6 and induced with 1mM IPTG for 4 hours.
All the constructs expressed protein at the expected size.
L=ladder, U=uninduced, I=induced
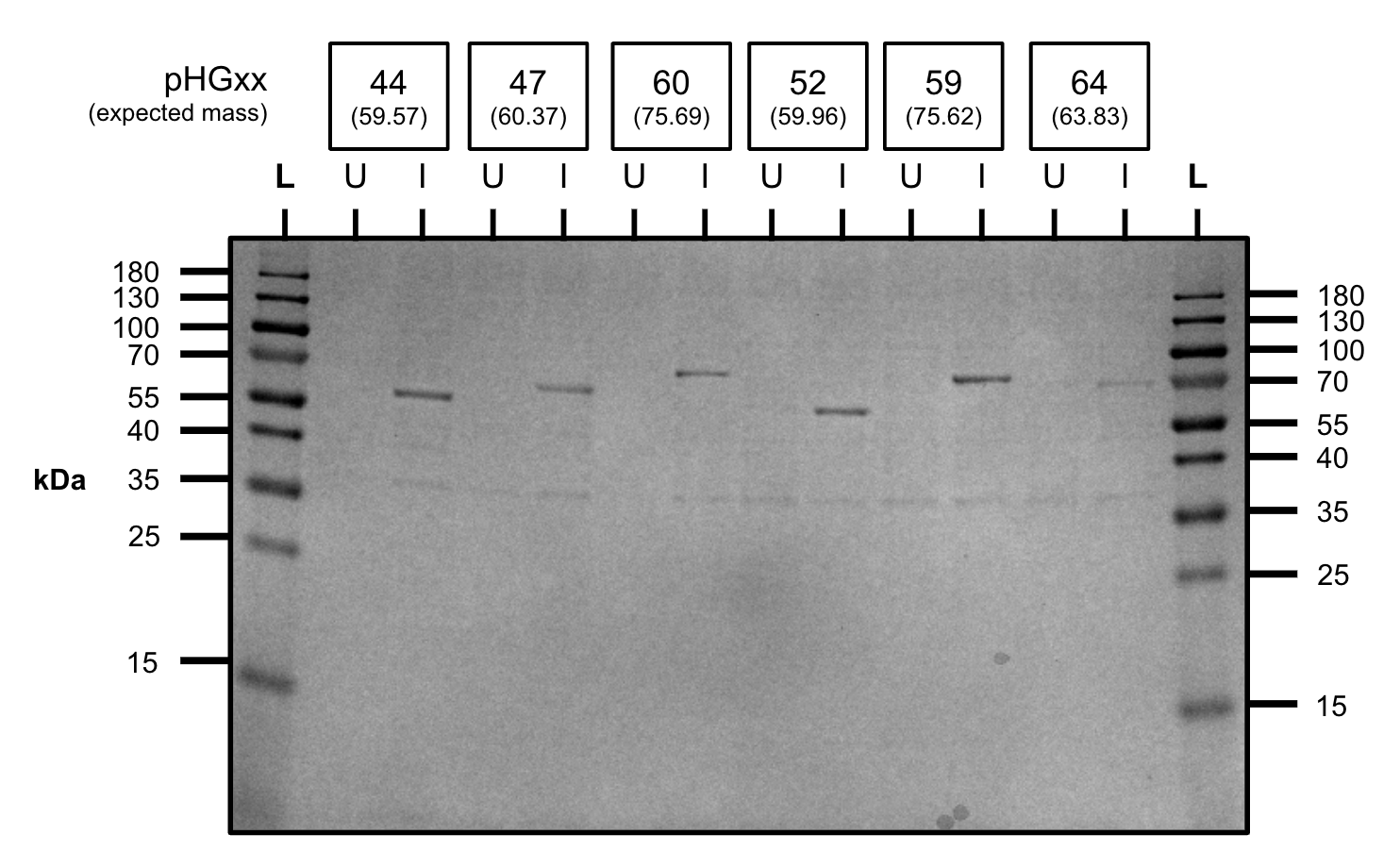
Expression test (SDS-PAGE)
Expression test (SDS-PAGE)
Large scale expression of pVS23 for lysate mixture tests.
We received a his-stain that can be applied to SDS-PAGE gels to visualize His-tagged proteins.
We tested the stain on a lysate mixture of pVS22 and pVS23.
Unfortunately we were unable to get access to the gel imager over the weekend and the gel destained considerably leaving faint bands.
Lysate mixture of pVS22 and pVS23 (Coomassie brilliant blue) (SDS-PAGE)
Lysate mixture of pVS22 and pVS23 (InVision His-tag stain) (SDS-PAGE)
Ran colony PCR screening on several sspDnaB constructs from the 2019 iGEM distribution kit.
Week 22
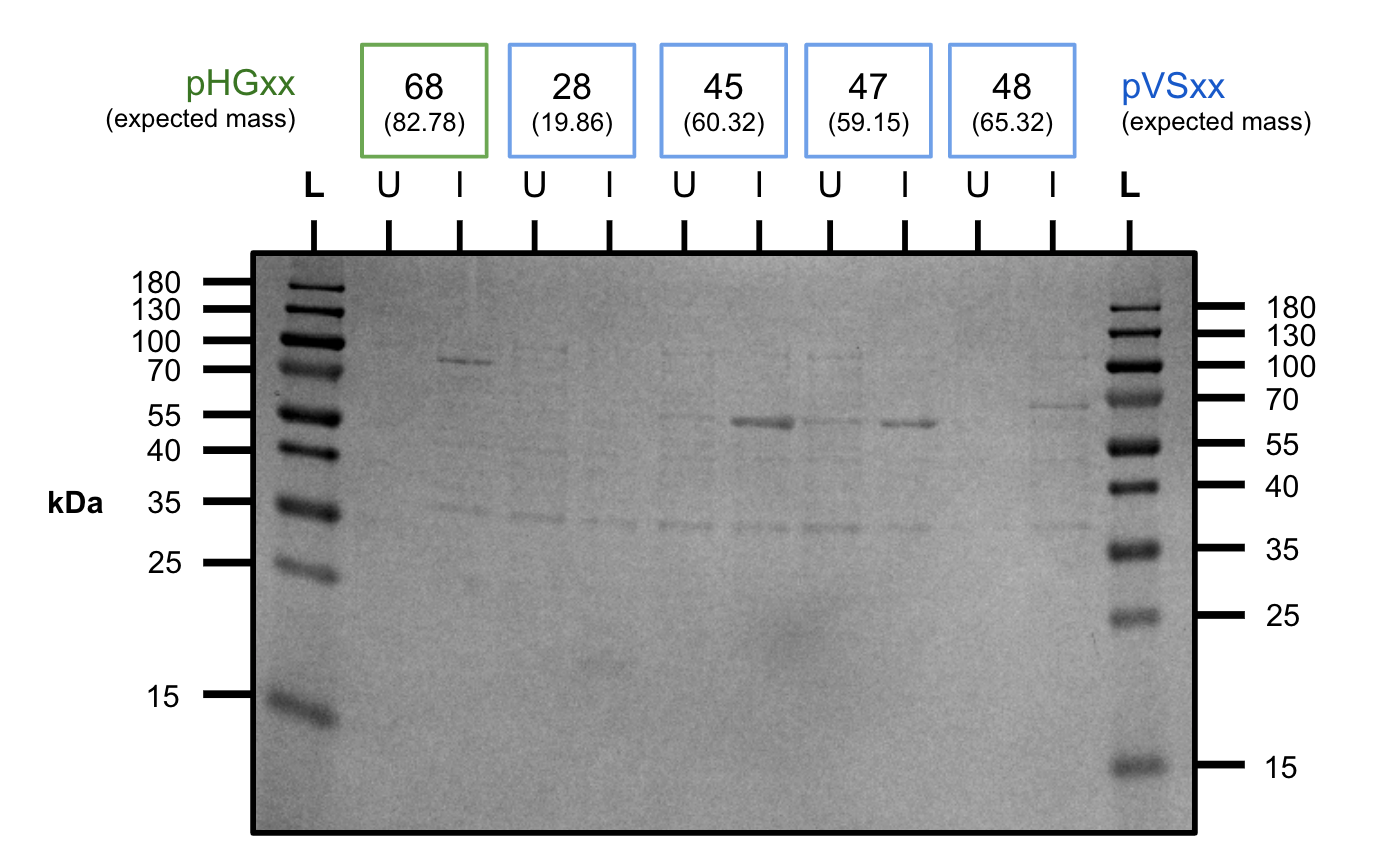
Tried expressing pHG68 at room temperature and 37 C to see if it would automatically splice like a cis intein.
While the construct expressed at the right size, we did not observe band formation corresponding to the spliced product.
On this gel we also tested diafiltration of pVS30 (E=elution, Df=after buffer exchange) to ensure our protein was retained during buffer exchange.
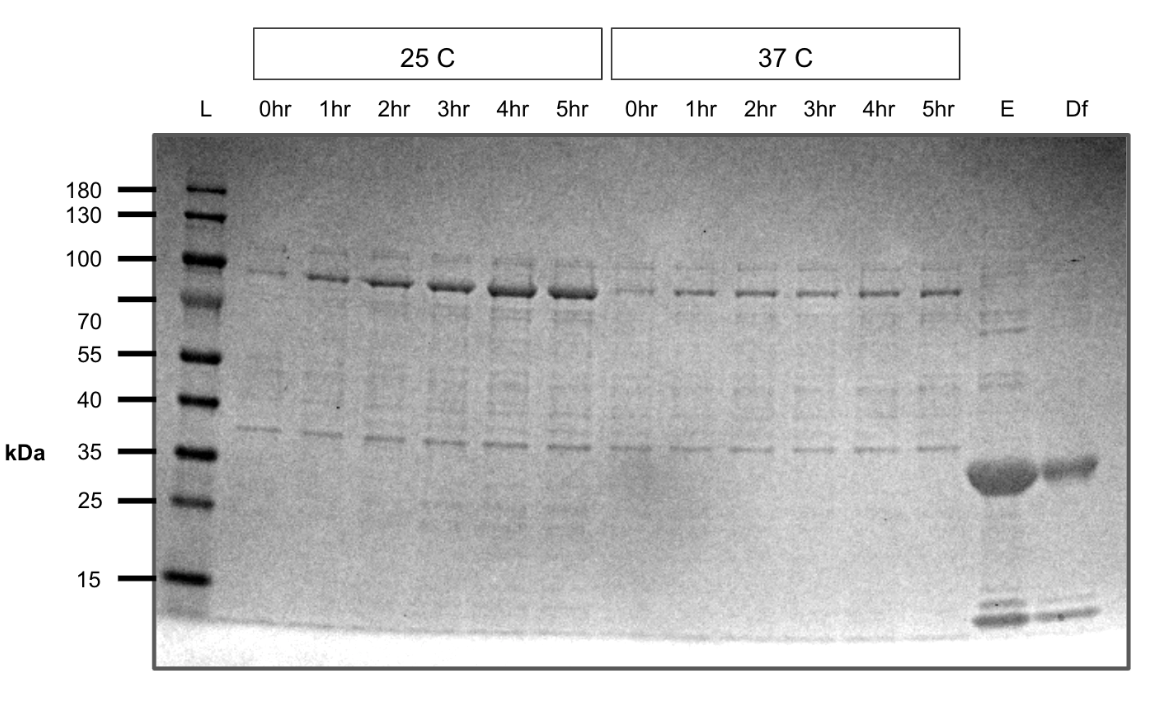
pHG68 expression test (SDS-PAGE)
Grew 200mL cultures of pHG(44,52,60,64) to OD600=0.8-0.9 and induced with 1mM IPTG for 4 hours. Cultures were pelleted down and stored in -80 C for purification.
Tested the new diafiltration columns on pVS29 to concentrate and exchange buffers.
Large scale expression of pVS47 for lysate mixture tests.
Redid Gibson assemblies of constructs that failed colony PCR screening from the third nested intein set and the split-linker set.
Purified 200mL cultures of pHG(44,52,60,64) from frozen pellets.
All contructs except for pHG60 purified well.
pHG44 also has a relatively thick band in the elution below the expected size which we believe is a degredation product containing maltose binding protein.
U=uninduced, I=induced, P=pellet, S=supernatant, Ft=flow-through, W=wash, E=elution
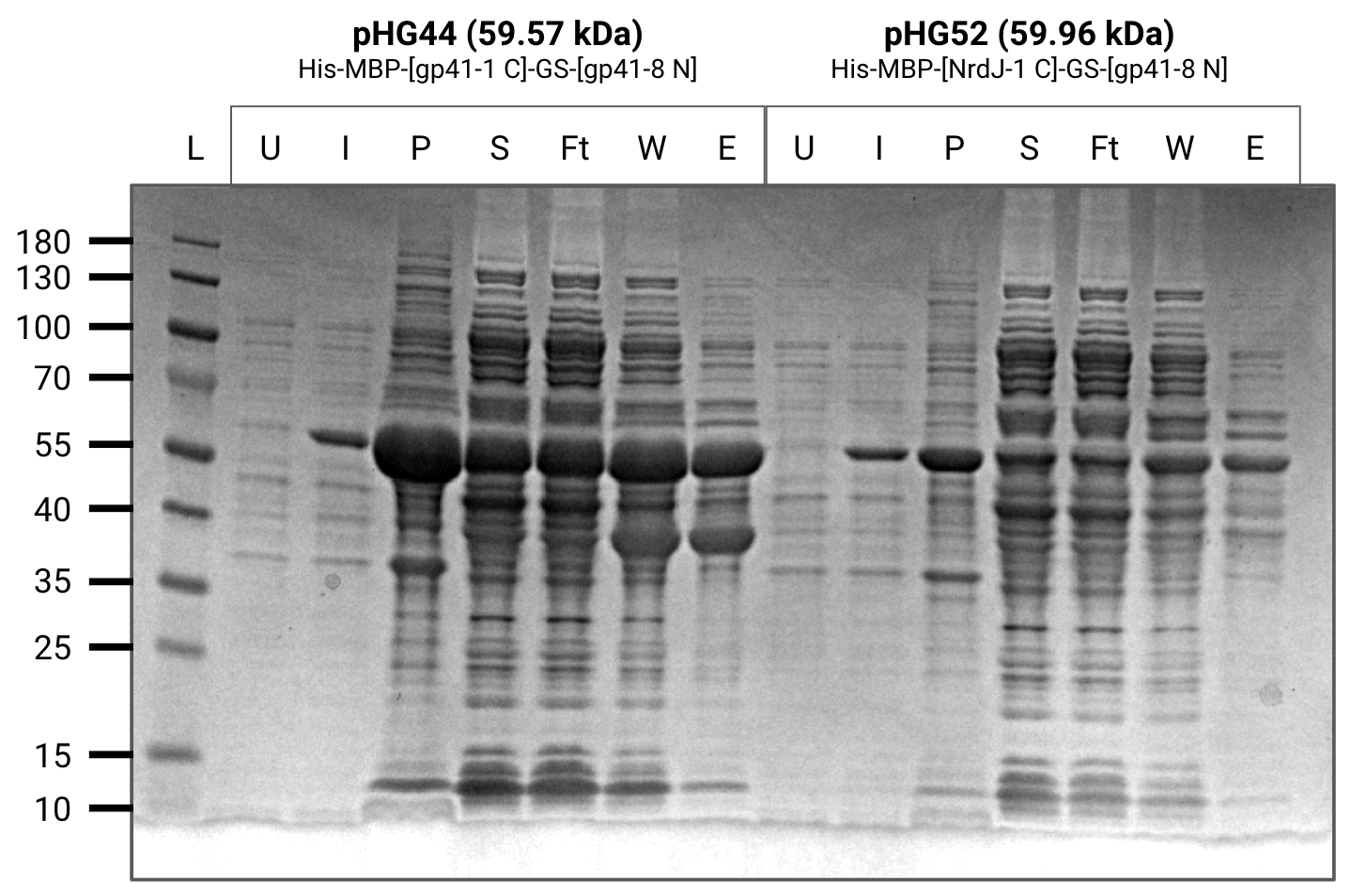
Purification gel (SDS-PAGE)
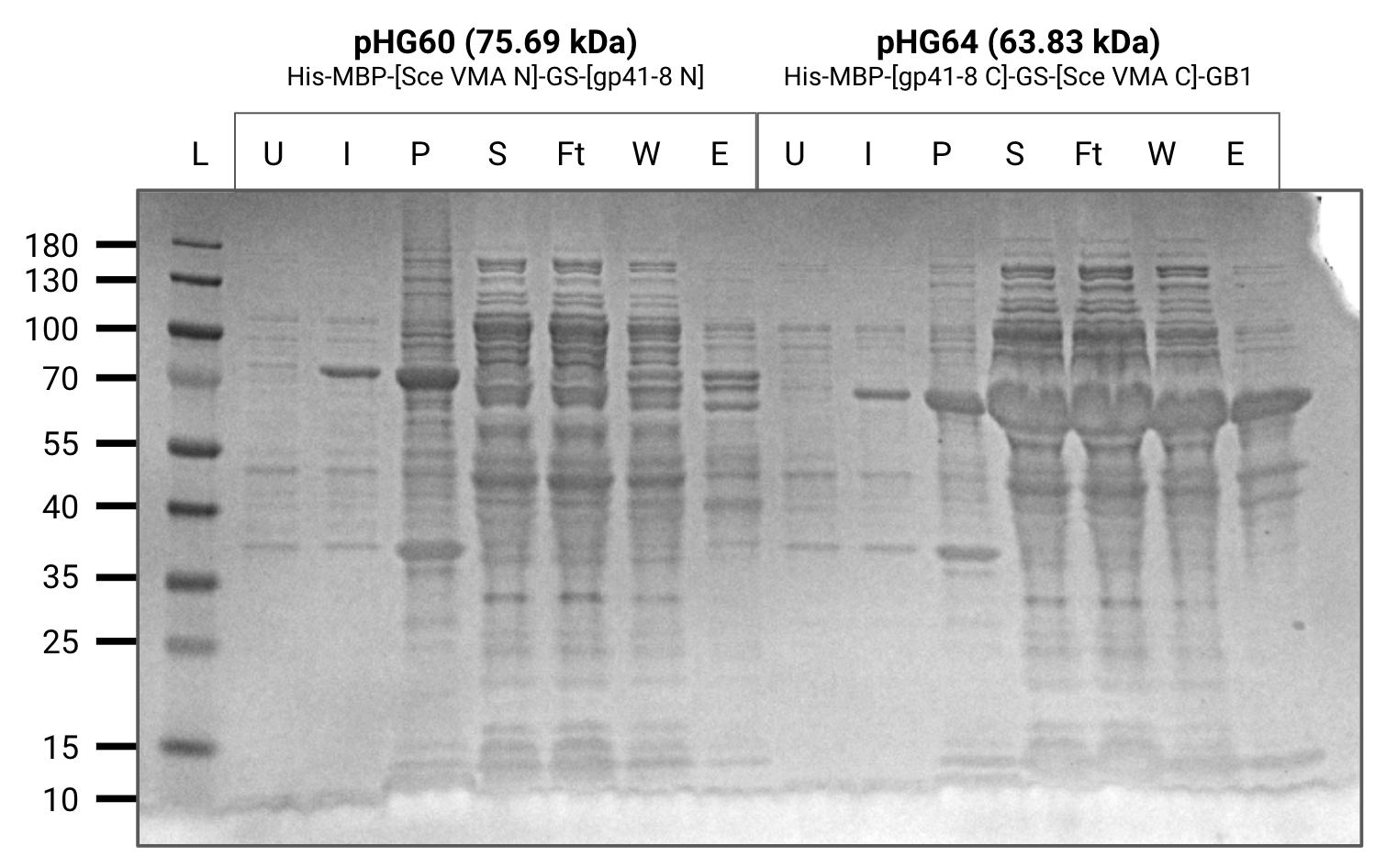
Purification gel (SDS-PAGE)
Large scale expression of pVS48 followed by purification and diafiltration.
Elutions of the four constructs were concentrated in centrifugal diafiltration columns and resuspended in a splicing buffer with high glycerol content intened to mimick the intracellular environment.
Several of the constructs were mixed in pairwise in-vitro assays to see if splicing would occur.
We were able to observe splicing between pHG44 and pHG64 however the concentration of pHG52 and pHG60 was too low to show up clearly on a gel.
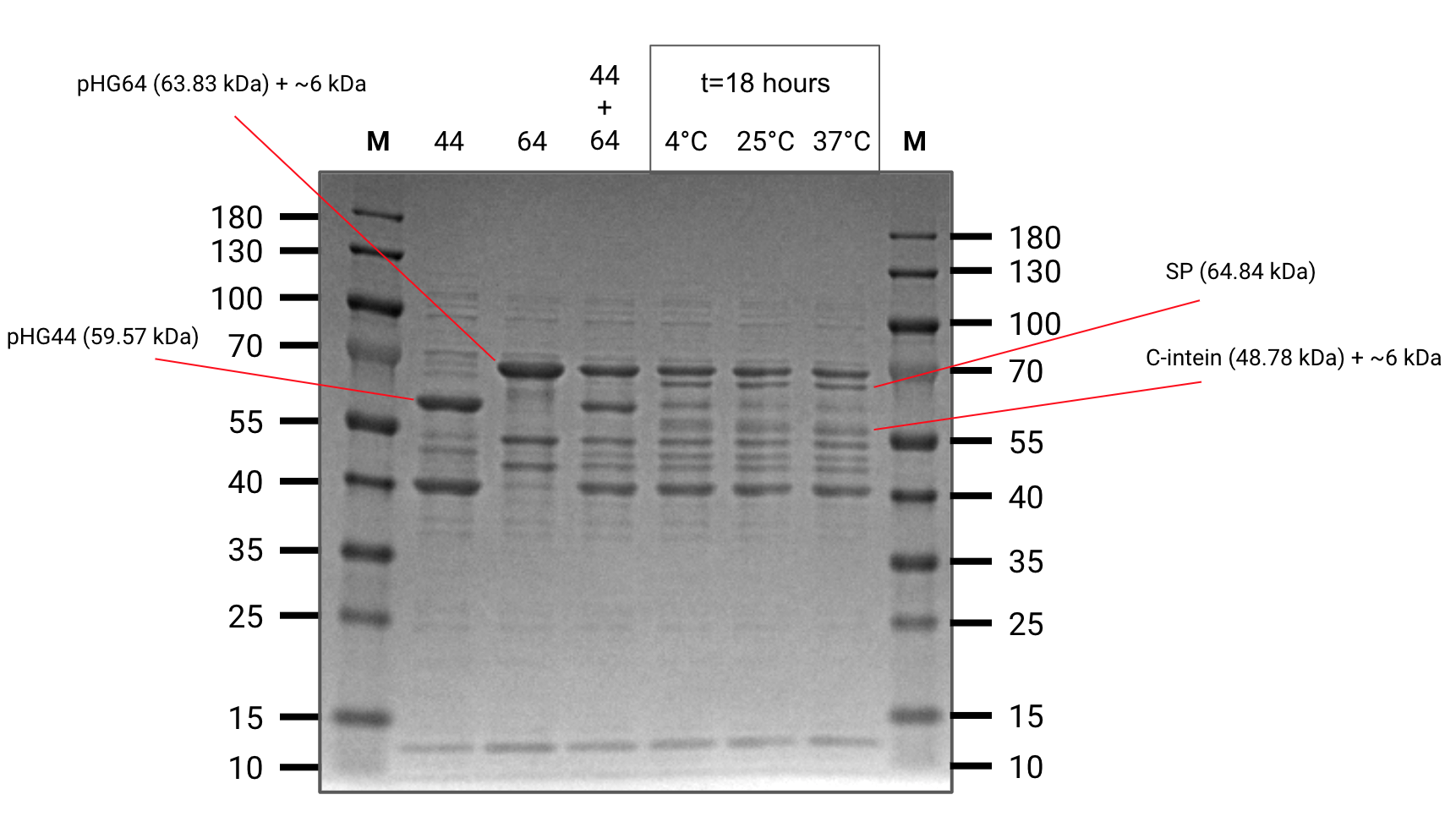
Splicing assay (SDS-PAGE)
Important reactants and products are annotated
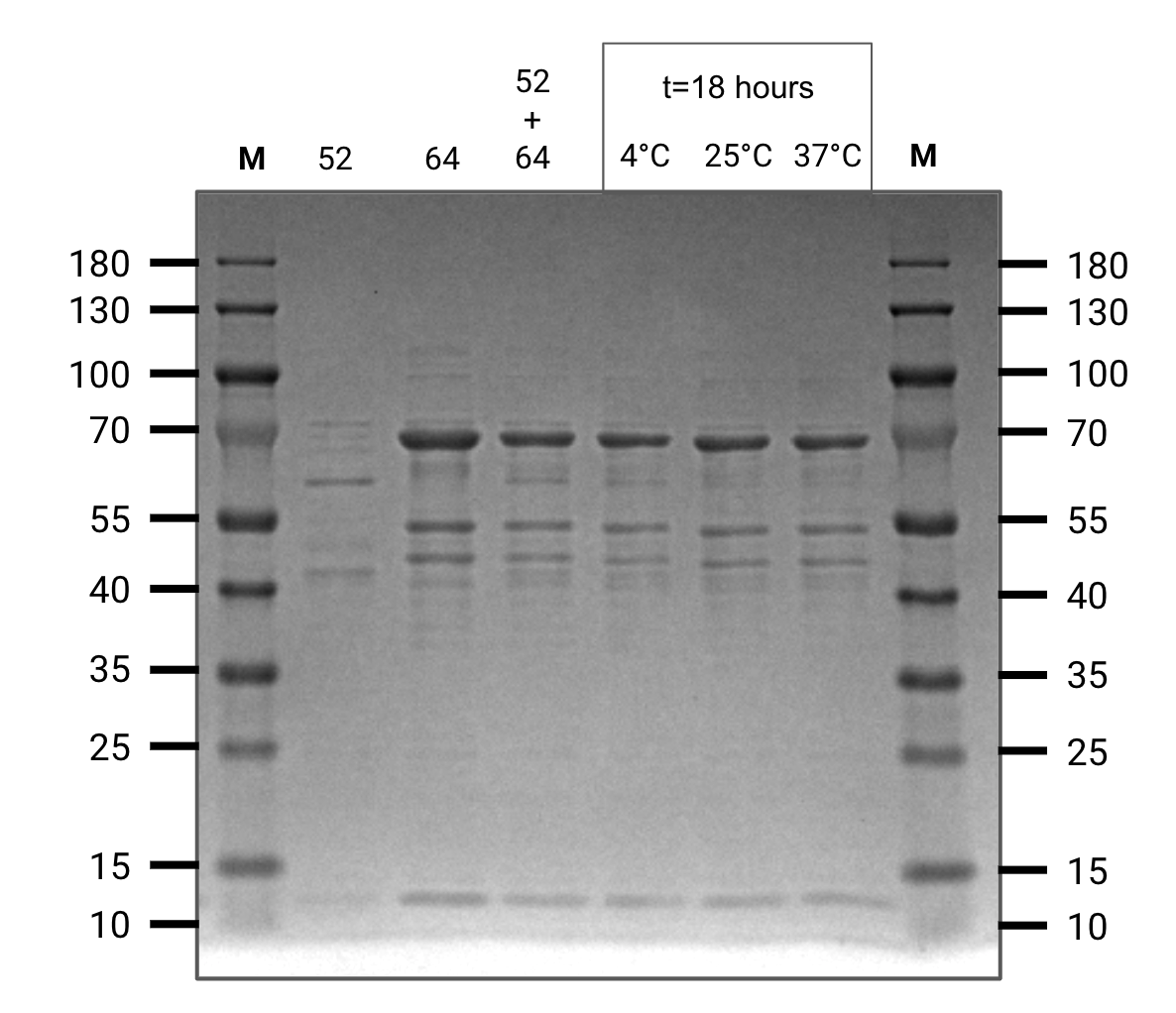
Splicing assay (SDS-PAGE)

Splicing assay (SDS-PAGE)
Grew 200mL cultures of pHG(44,59,60,68) to OD600=0.7-0.8 and induced with 1mM IPTG at room temperature for 4 hours.
Cells were pelleted and stored in -80 C for purification.
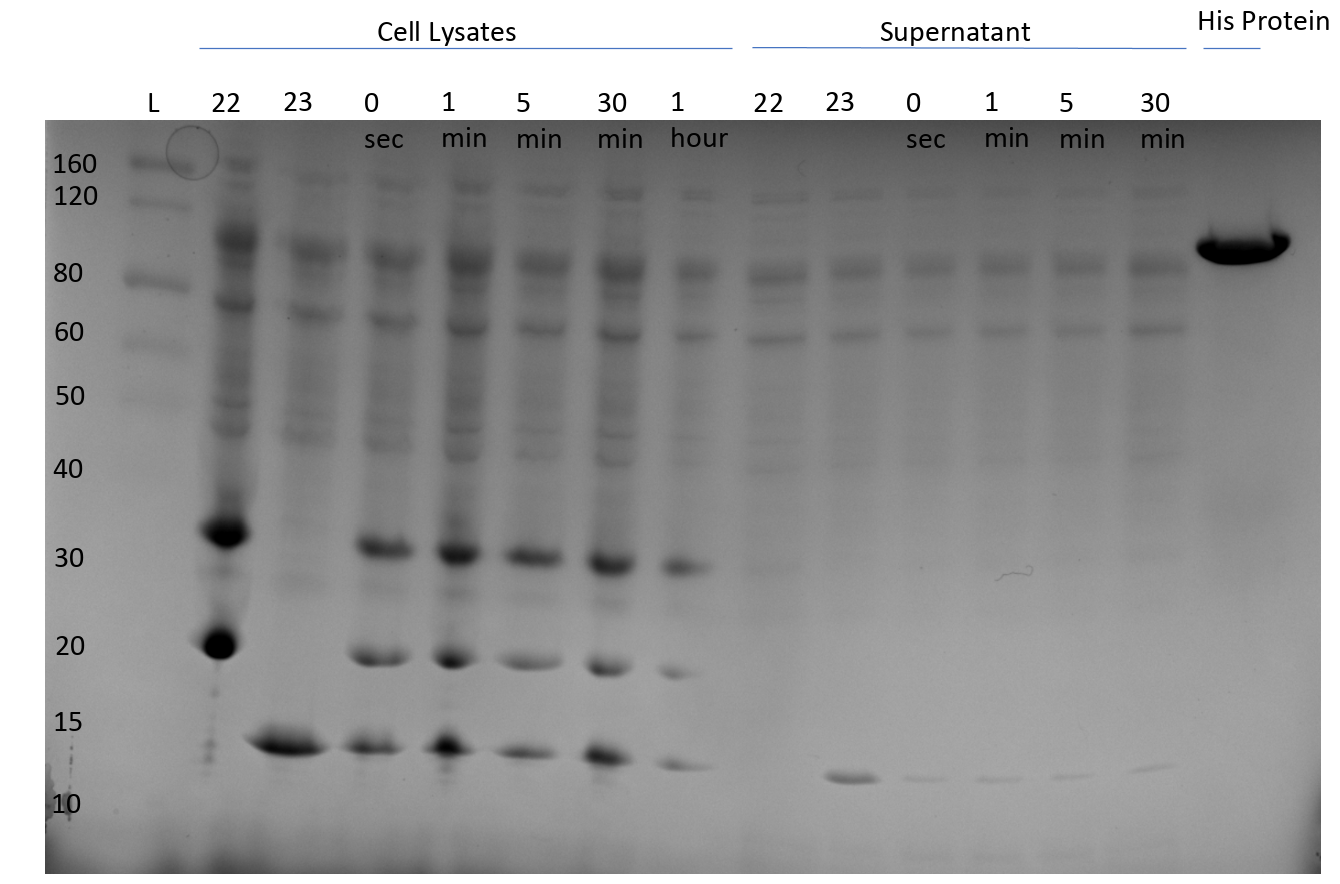
After observing splicing between pHG44 and pHG64, we ran another gel and took aliquots at certain time intervals to develop an understanding of the reaction rate.
We also ran both starting constructs at t=4 hours as negative controls to ensure we were not seeing degredation products.
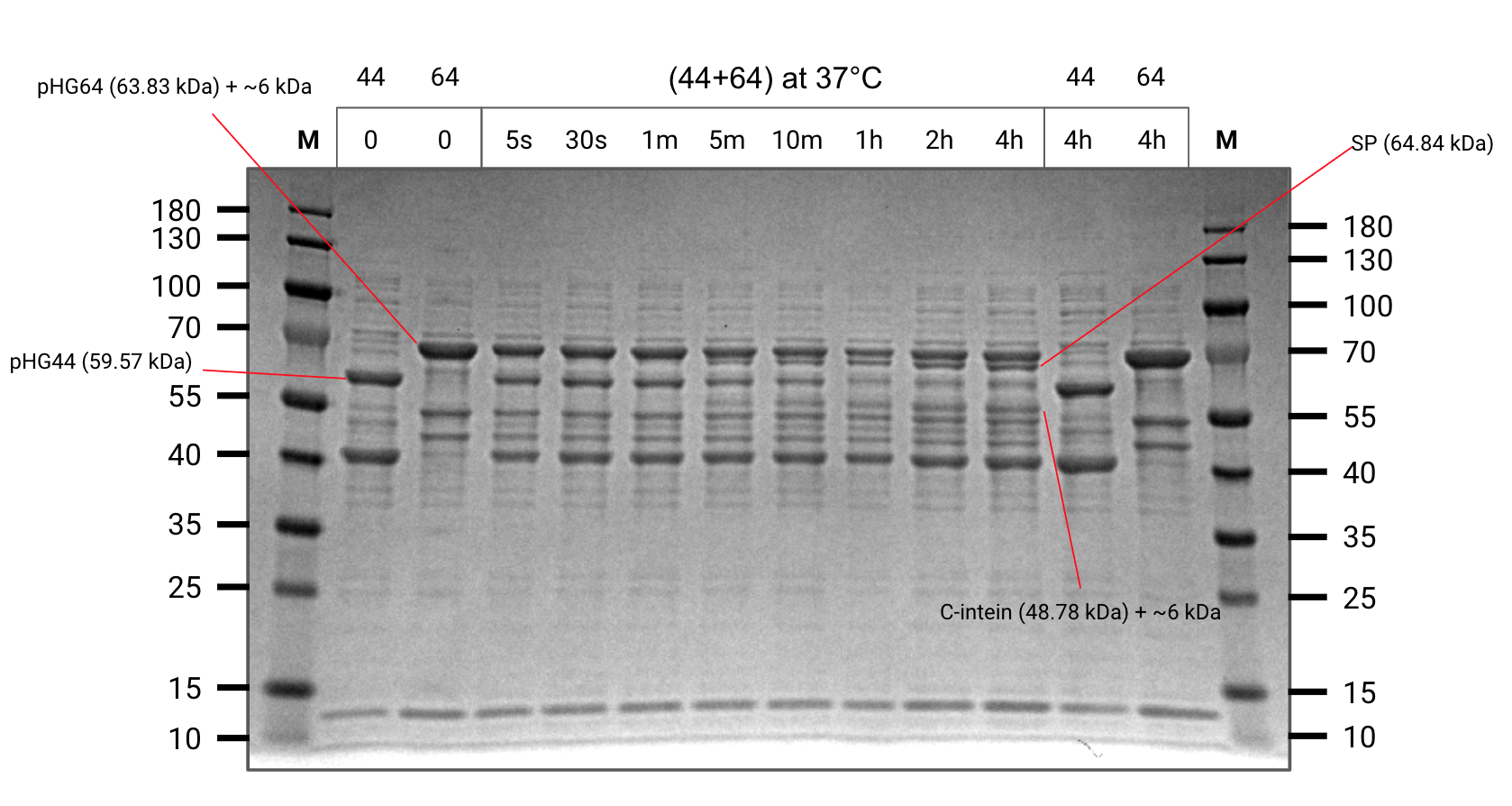
Splicing kinetic assay (SDS-PAGE)
Important reactants and products are annotated
Transformed a construct from the 2019 iGEM distribution kit for characterization.
Week 23
Large scale expression of pVS47 and pVS48 for lysate mixture tests.
Purified 200mL cultures of pHG(44,59,60,68). All constructs purified well.
U=uninduced, I=induced, P=pellet, S=supernatant, Ft=flow-through, W=wash, E=elution
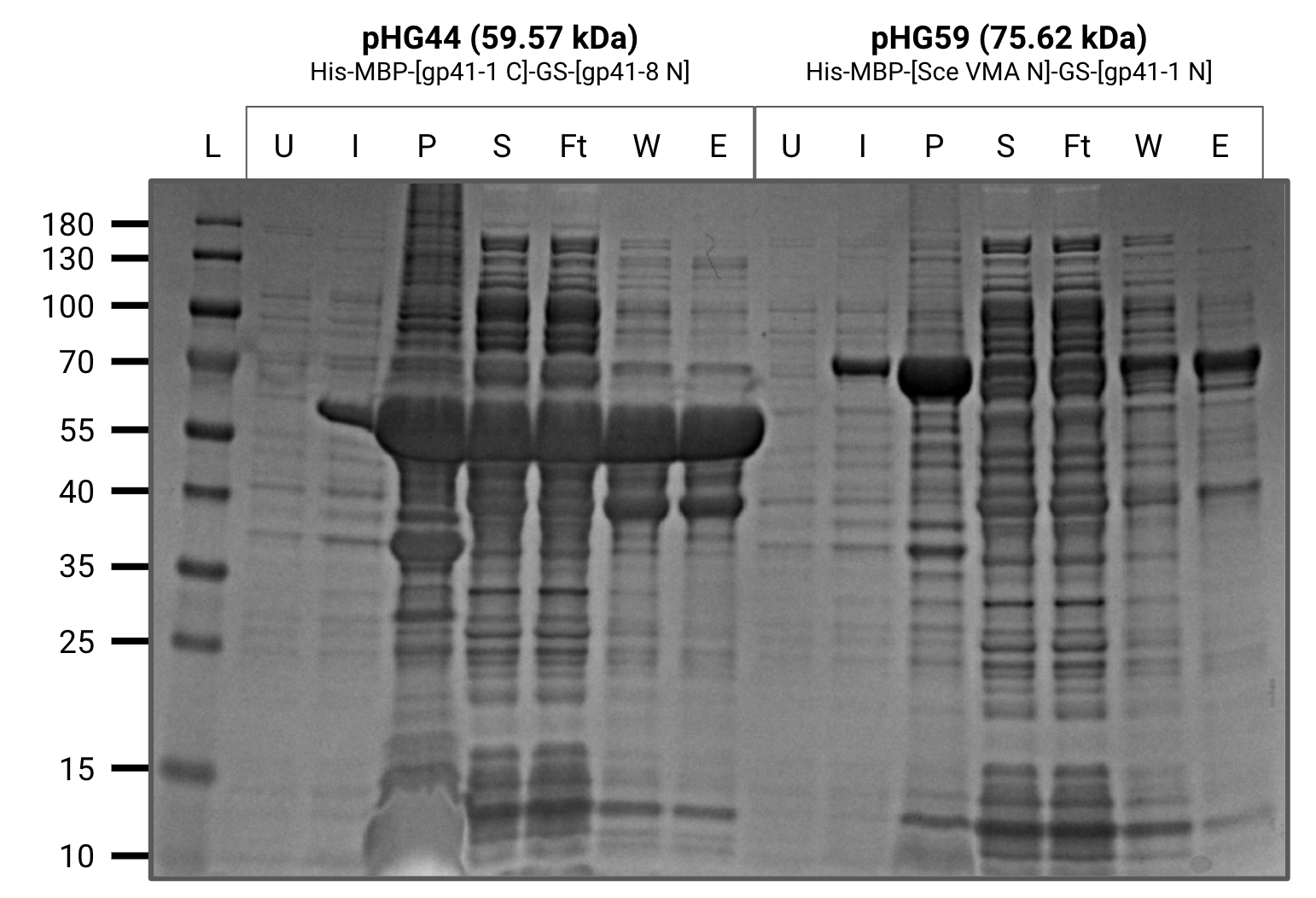
Purification gel (SDS-PAGE)

Purification gel (SDS-PAGE)
Large scale expression of pVS45 and pVS29 for lysate mixture tests.
Performed overnight in-vitro assays of several combinations of the recently purified constructs.
We observed splicing between pHG44 and pHG64 (as seen before) and also a hint of splicing between pHG60 and pHG64 (predicted).
However, we did not observe splicing between pHG59 and pHG44 as expected. After carefully reviewing our construct sequences, we realized that our design was missing a single amino acid residue in gp41-1 N.
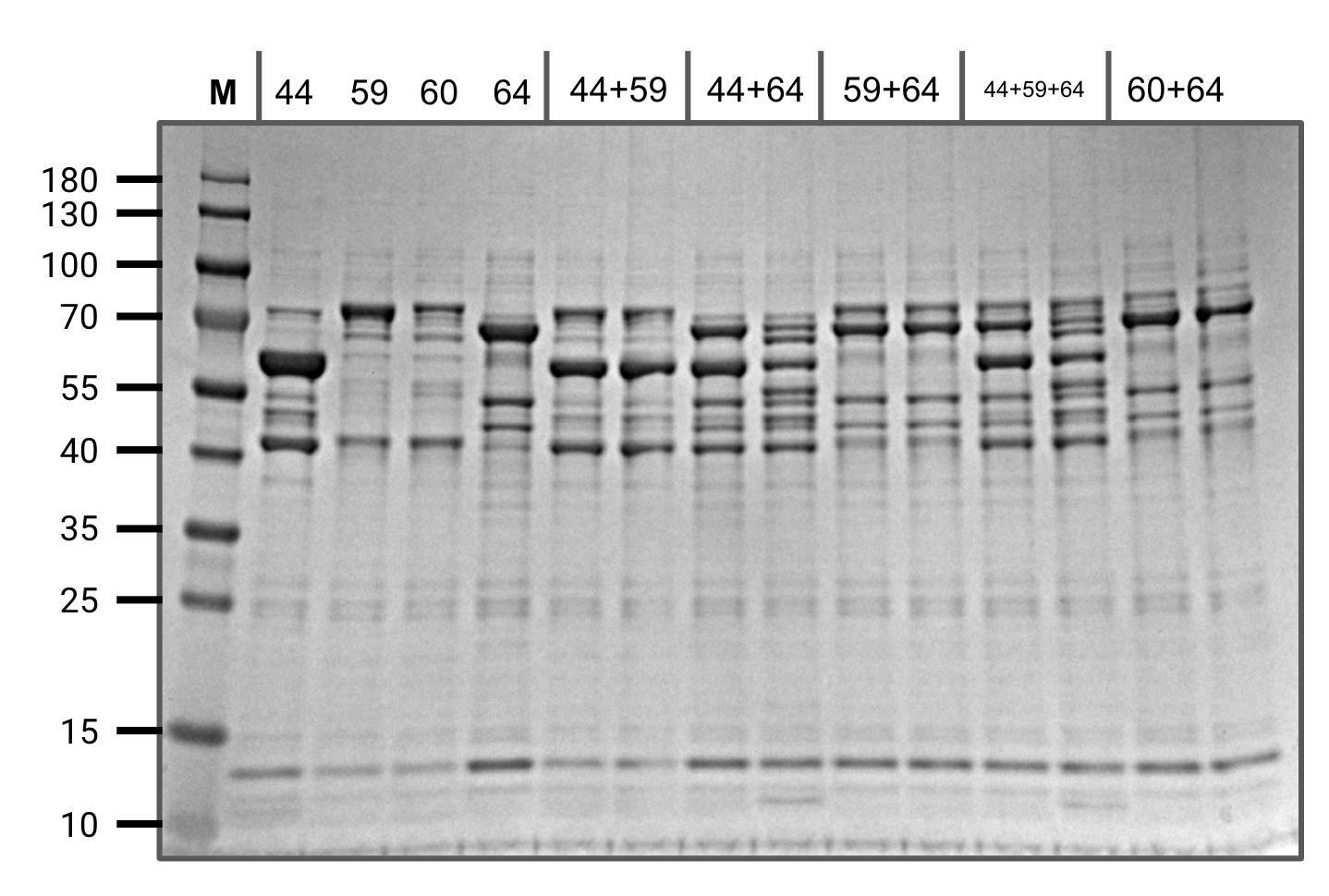
In-vitro assay screening (SDS-PAGE)
The four constructs on the left are at t=0
For each reaction mixture, the left lane is t=0 and the right lane is t=20 hours at 37 C
