Introduction
Our team wanted to explore the possibility of protein-based logic systems. Although protease systems have previously been designed, we wanted to explore the diversity of inteins in order to implement this post-translational logic. Previously, three-input AND gates were designed by splitting the extein sequence into three parts [Lienert 2013]. We wanted to design a system that would allow for minimal disruption and scarring of the extein sequence and a system that would allow for the cascading of splicing events. As a result, we designed a two systems to achieve these goals: "nested inteins" and "split-linkers".
We first designed the "nested intein" system in which a trans-splicing intein disrupts a second trans-splicing intein's N or C terminal. In this setup, disruption of the intein terminal would render it nonfunctional. Thus, the first splicing event would result in the ligation of a full intein terminal, leaving it functional. As a result, this system would allow us to cascade splicing events so each splicing reaction directly impacts the next.
Alternatively, we designed our "split-linker" system. In previous literature, the SceVMA intein was characterized as weakly associating intein, likely because this intein is artificially split. Building upon studies that used protein scaffolds to increase the effective concentration of SceVMA in solution [Selgrade 2013], we designed a system in which the SceVMA intein was connected by a GS linker that was split by orthogonal inteins. This meant that splicing events would result in the creation of the full GS linker.
Both of these systems allow for versatility in designing logic gates and proposing the possibility of intein-based cellular circuits.
When deciding which inteins to pick for our project, we took into consideration kinetics of the splicing reactions and orthogonality of the inteins. The optimal splicing conditions outside of the junction sequences all had to be relatively identical to each other, since the inteins would exist in the same construct together. This included pH and temperature optimization of the reaction conditions. Once we found inteins that fit these parameters, we then considered split sites to determine which inteins were the most compatible with one another.
The inteins we chose were Npu DnaE, gp41-1, gp41-8, IMPDH, NRDJ-1, and TvoVMA. We chose these inteins because of the previous literature that cited these inteins as both kinetically fast and orthogonal to each other [Shah 2012] [Shah 2014]. In our split linker constructs, we also included SceVMA, since studies suggest that this intein is weakly associating, requiring a system to increase the effective concentration in solution.
Nested Inteins
Design of each protein or protein chimera which would utilize the nested intein system requires a top-down approach. First, the inteins that does the final splicing has to be chosen. Next, additional intein pairs must be chosen which would disrupt functionality.
When picking the split sites, we considered the secondary structure of the site, the native junction sequence of the intein, and the block regions of the intein. We looked for the structure of the site to be away from the host intein's block regions and were not right in the middle of a crystal structure. We preferred places that were in turns or disordered regions according homology models. We did this to help prevent misfolding of the host intein after the nested spliced out. The native junction sequence was chosen with a set of rules. The most important one being that the C+1 junction was a match to the native sequence. The C+3, N-2, and N-3 sites were lower priority since available sites were limited and were not considered to affect the splicing rate as much as the N-1 and C+2 sites. Whether we could change the N-1 and C+2 sites was answered based on our own theories and known data of a few inteins. If we felt the site could not be changed easily we looked at the native amino acids in these regions and looked for ones with similar structure. For example, if the C+2 site was tyrosine we would look to see if the replacement amino acid had resonance and hinderance.
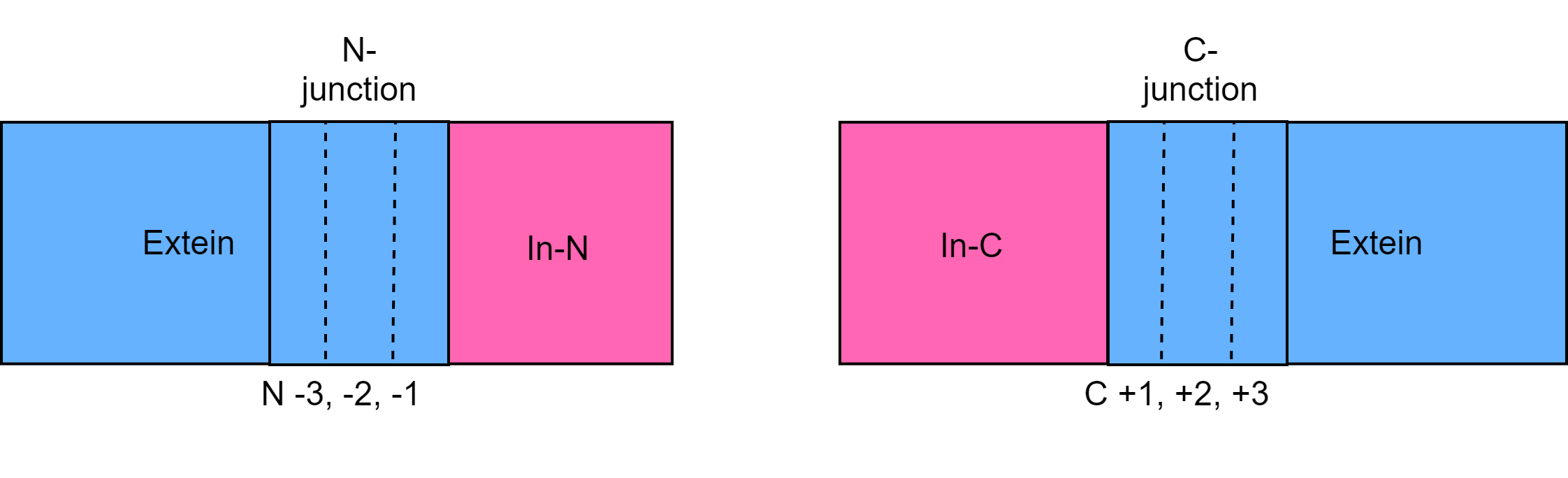
To learn more about the chemistry of Inteins and how we chose split sites, click here!
To choose the insertion sites of our inteins, we developed a prototype splice site calculator which analyzed intein sequences for potential splice sites. This calculator prioritized that the C1 site matched the native sequence, then ranked how well the N1 and C2 junction sequences matched the native sites, and gave a prediction of the secondary structure at the split site. The predictions were made using the BLOSUM amino acid substitution matrix. With this calculator hundreds of split sites were filtered out and the ones left we were able to manually go through them and pick the ones we believed would work best.
Furthermore, we took the highest ranked predicted splice sites and compared it to the crystal structure or homology model of the protein. Splice sites located in areas of secondary structure were not considered due to high possibility of disrupting block sequences (more information on block sequences can be found here). We aimed for splice sites located in loops or disordered regions.
Nested Intein Optimization
After consulting with Dr. Hideo Iwai from the University of Helsinki, Dr. Henning Mootz from the University of Munster, and Dr. Seth Horne from the University of Pittsburgh, we decided to find additional splice sites where we could make a point mutation in the flanking region. The goal of the mutation was to increase splicing efficiency.
All of our constructs were designed for either pTEV5 or pET28a vectors which contains antibiotic resistance, an IPTG inducible promoter, an N-terminal 6x His-tag, and a protease site for His-tag removal. Our choice behind which vector to use was dependent on which gene synthesis company we ordered our genes from (pTEV5 constructs were from IDT and pET28a constructs were from Twist Bioscience).
Once we began our protein expressions, we found our proteins were highly insoluble and aggregating our pellet. We consulted with PhD candidate Dylan Tomares from Dr. W. Seth Childers' Lab at the University of Pittsburgh who suggested incorporating a solubility tag into our protein constructs. In response to this, we re-cloned one set of constructs into the pTEV6 which contains a N-terminal maltose binding protein between the 6x His-tag and the protease site .
Split-linker System
In designing our split-linker constructs, we made similar considerations about junctions sequence dependency, kinetics of the reaction, and orthogonality of the inteins. However, the advantage to this system was the fact that the junction sequences could be hidden within the linker region. This allows us to completely preserve flanking sequences for the intein and also prevent additional amino acid residues to be added to the extein sequence.
Since our flexible linker region consisted of "GGGS" sequence repeated three times. Embedded within this repeated linker sequence are the native junction sequences and the N and C terminals of inteins. This allowed us to create modular inputs for each of the reactions. Both three-part and four-part ligation systems were designed and these constructs can be seen in the figure to the right.
Due to the issues that we encountered with solubility in our nested intein constructs, we immediately cloned these inserts into pTEV6 with maltose-binding protein in order to attempt to circumvent these issues.
Future Work
We will continue working on this project by completing cloning, expression, and purification of our other sets and constructs in order to continue to provide evidence for of intein-based cellular circuits. We hope to perform assays with the construct in which we introduced a substitution in order to determine if splicing efficiency was affected. We also hope to continue working on performing assays in order to better determine how the kinetics of these splicing reactions are affected by the systems that we designed.
References
Lienert, F., Torella, J. P., Chen, J. H., Norsworthy, M., Richardson, R. R., & Silver, P. A. (2013). Two-and three-input TALE-based and logic computation in embryonic stem cells. Nucleic Acids Research, 41(21), 9967-9975. https://doi.org/10.1093/nar/gkt758
Selgrade, D. F., Lohmueller, J. J., Lienert, F., & Silver, P. A. (2013). Protein scaffold-activated protein trans-splicing in mammalian cells. Journal of the American Chemical Society, 135(20), 7713-7719. https://doi.org/10.1021/ja401689b
Shah, N. H., Dann, G. P., Vila-Perelló, M., Liu, Z., & Muir, T. W. (2012). Ultrafast protein splicing is common among cyanobacterial split inteins: Implications for protein engineering. Journal of the American Chemical Society, 134(28), 11338-11341. https://doi.org/10.1021/ja303226x
Shah, N. H., & Muir, T. W. (2014). Inteins: Nature's gift to protein chemists. Chemical Science, 5(2), 446-461. https://doi.org/10.1039/c3sc52951g
