Overview
While the optimal iGEM team size is 8 to 12 members, the average team contains only 5 full-time equivalents during the summer research months. We all know why this is—college students are busy. Whether it’s a limitation on volunteer hours or it’s a cap on the number of stipends a university can offer, the reality is that most teams are left wanting more person-power at the end of the day.
Another reality is that there are thousands of high school students that have had exposure to the fundamentals of molecular biology through classes, but who are still hoping to gain experience working with hands-on research. In fact, a recent survey showed that 77% of high schoolers “are either extremely or very interested in volunteering to gain work experience.”
Seems like these two realities could perhaps combine synergistically?
……Our team thought so too.

DiCE in vivo
Experiment 1: Does Qβ Self-Replicate in E. coli?
To summarize this first experiment, we tested for the ability of Qβ replicase to replicate sense mRNA, transcribed off a plasmid vector. In order to do so, we tested for the antisense strand of RNA, which could only form as a result of replication with Qβ. This was accomplished by, in short, synthesizing cDNA from extracted RNA after one 12-hour growth phase in a chloramphenicol dosage assay. Using primers, specific to either sense or antisense RNA strands to synthesize the cDNA, we could detect the presence of antisense RNA by running gels of both antisense and sense cDNA.
It is important to note that this experiment was run with the initial construct design, pRNA, as described in the design section above. For simplicity, the target gene is shown in green while the gene encoding for Qβ replicase is omitted as it is not necessary to mention for this experiment. The major steps taken for this experiment are depicted below.



Experiment 2: Can Qβ replicase Increase Antibiotic Resistance?
While it does not substantiate the hypothesized mechanism depicted in the design section, it does suggest to our team that perfecting the DiCE system is possible within E. coli. It is important to note, however, that this experiment was done using pRNA, and not the optimized constructs. A more comprehensive set of experiments with the new construct designs, No Qβ and Qβ have been designed and are awaiting the success of our mRNA gene delivery protocol for E. coli.
To summarize the experiment, several rounds of chloramphenicol dosage assays were done to provide an increasing selective pressure to increase resistance by optimizing the gene (again, this was using the pRNA construct and, therefore, can’t substantiate this hypothesis without sequencing data). After each round, cells that had survived in the highest concentration of chloramphenicol were moved into the next assay at even higher concentrations.
Experiment 3: Can mRNA Gene Delivery Be Accomplished in E. Coli and Maintained Qβ replicase?
This experiment is still being troubleshooted due to the overwhelming lack of literature on the subject of mRNA gene deliver in E. coli. Currently, this experiment is being attempted under several varying conditions and is also awaiting new reagents and parts. It is crucial to note that this experiment is utilizing the new construct designs, No Qβ and Qβ. Our protocol for creating electrocompetent cells can be found here in a Google doc as it is still under experimentation and not ready for publication on our website. Additionally, our electroporation protocol can be found here. Conditions tested are shown below:

DiCE in vitro
This section contains both resources for and details on the implementation of the following internship program components:
- Program Outreach — Spreading the word
- Application Process
- Recommendations for Gaining Departmental Support
- Safety
- Mentorship Structure
- Gathering and Implementing Program Feedback
- Closing Out the Program
Program Outreach
A common pitfall of outreach is to selectively reach out to those who are already well-connected to such opportunities. For our team, being situated in the Bay Area made this all the more pertinent: it would be all too easy to get ample applicants by simply contacting one or two private high schools with already well-established internship programs. We wanted to reach beyond this, however. Thus, we made it our goal to spread the word to people who may not otherwise have the opportunity to get engaged with lab research.

- Had each Stanford iGEM member contact 2-3 local biology teachers from a variety of high schools.
- Had a departmental-wide announcement to graduate students advertising the program and encouraging them to forward our program information widely.
- Did in-person outreach to spread the word to local high school students.
We put together and distributed the following informational packet to these different outlets. Note that this is a version prior to the expansion of the program to include minors: https://tinyurl.com/Stanford-iGEM2019-18program
More recently, we improved the cover page to the following (see right). An editable version of this template can be downloaded here for teams interested in building upon this handout to use for their teams

Application Process
The goal of the application process was to identify candidates that have demonstrated interest in pursuing science beyond the classroom and that would be committed to the team for the duration of their internships. We emphasized that prior lab experience was not required or expected, and we encourage applicants to highlight other skills and experiences outside of the classroom that may have given them applicable skills to working with a team on a long-term project.
We designed an application process with two phases: a written application with a letter of recommendation from a teacher or mentor, followed by a face-to-face interview with two to three members of the iGEM team.
For the application we asked for a one-page cover letter addressing the following four questions:
- Why would you like to work on the Stanford iGEM team this summer?
- What level of commitment can you have this summer?
- What prior experiences have you had that may be relevant to your experience this summer?
- How have your prior experiences uniquely prepared you to contribute to iGEM?
They could submit this cover letter along with basic information at this google form. As an additional resource, here are email templates that our team used throughout the process. Although teams may need to modify these documents to to tailor them for their respective programs, we encourage teams to use them as starting points for their program and to reach out to us if they have any questions.
Gaining Departmental Support
Having minors work supervised exclusively by undergraduates in a lab with a variety of safety hazards creates significant liability. Thus, getting support for a program such as this is inherently political and can be a time-intensive and bureaucratic process—get started well-before summer to leave time for outreach and a substantive application process!
Our first recommendation is to reach out to immediate mentors that know the larger system within which your iGEM team is operating. For us, this was the teaching lab’s Lab Manager. She knew what entity would ultimately need to approve such a program (the BioE Department Chair), she had recommendations for putting together a proposal, she knew some of the safety training requirements and pointed us to resources for minor-specific requirements, and finally she could connect us to the intermediate faculty from which we would need to garner support before proceeding to the department chair.
Our final proposal was a modified version of the aforementioned informational handout, but with an additional letter of support from our lab manager.

My final more personal recommendation here is to balance being both professional and personal as you build relationships with those across the department. Though it may at times seem like a bureaucratic web of rules, building mutual trust with all those you come across can go much further than you may expect.
Safety
Safety was at the forefront, tail-end, and just about every step of the internship program. Before entering lab we had a comprehensive list of required safety trainings and documentation to complete, as well as a detailed list of requirements while working in lab.
In addition to the usual lab safety standards, working with minors poses a few additional hurdles. While these are unique for each university, beyond parental consent and liability forms from parental guardians Stanford also required that all of the undergraduates on our team completed additional “Working with Minors” online trainings, got official background checks, and were fingerprinted.
In addition to the safety checklist, we documented the expected types of lab work in which high school students may participate, and also outlined off-limit work (such as tissue culture and chemical room access). For all time in lab, we ensured high school students would always be paired with at least one undergraduate at all times.

Mentorship Structure
We approached the mentorship structure with intention: we aimed to create a program that efficiently used both undergraduate and high school students’ time, that would expose interns to a variety of types of work, and that would allow interns to take ownership over part of the project.
We encouraged interns to commit to 6-8 weeks (four week minimum), working in lab 4 hours per day Monday to Friday. Our team quickly realized that it was most effective to have the mornings reserved for the undergrads to do logistical work, long-term project planning, and initial lab work, then to have the interns join us in lab at 1:00PM for the remainder of the day.
We set aside the first 2 weeks for interns to shadow a variety of the sub-projects our team were working on in order to maximize exposure to different lab techniques. We put together a document with descriptions and links to protocols for common lab techniques (including basic instructions for using Benchling), and had interns print and annotate the procedures.

Following this initial phase, we then decided the interns were ready to take on their own sub-project: AcrIIA4 Directed Evolution. With assistance when needed, we let them design primers, submit sequencing data, and ultimately plan their own lab work timeline.
For the final phase we helped the interns write up the content of their sub-project for the website (check out “AcrIIA4 Evolution”!). While recognizing that research is never complete, this final push was critical for pulling together the work they had completed thus far, for recognizing and contextualizing the motivations for the work they were doing, and for ultimately sharing their findings with the larger scientific community.
Gathering and Implementing Program Feedback
From the get-go, we were up front with the interns that the program was a novel one. We highly encouraged open communication and constructive feedback on both positive and negative aspects of the experience so that we could continue to improve.
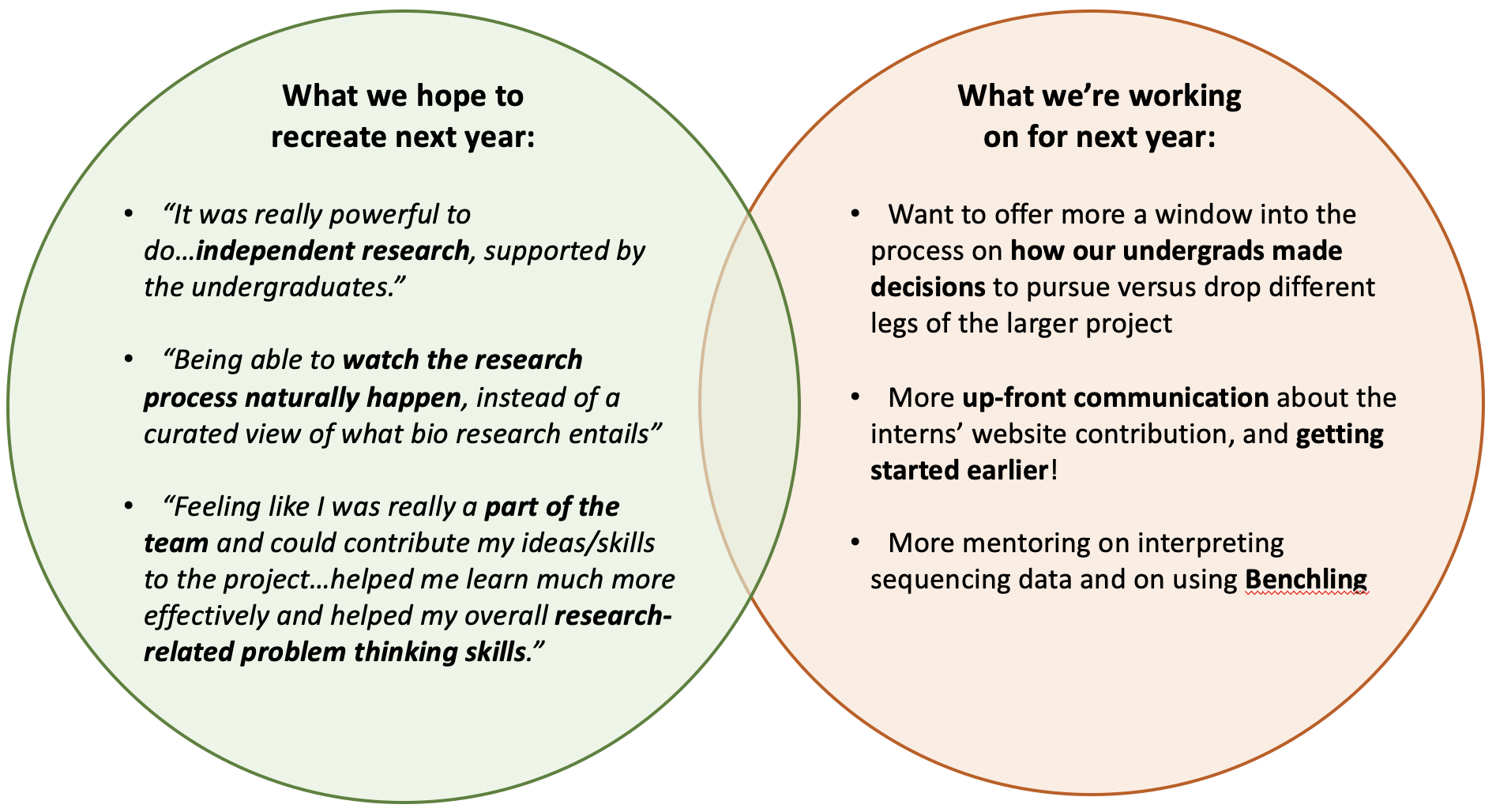
To further encourage and formalize this process, we had interns fill out an anonymous satisfaction and feedback survey both midway and at the end of the program. We included an input field for a random number the intern could put so we could track performance longitudinally while still maintaining anonymity. We tracked some metrics quantitatively such as overall satisfaction and whether the interns felt their time was being used effectively, and we also gave space for more qualitative feedback such as “What aspects of the program would you like continued” and “Please provide 1 to 3 areas where we could improve to maximize your experience this summer.”
These survey results both helped us to make tweaks to the program midway through the summer, and it also gave us concrete areas to work on for next year (see data below!).

Closing Out the Program
Though it may seem a small detail, we’re strong advocates of a fun last day :) On one of the last days of summer, we invited the three interns back for one last celebration of all the hard work we completed over the summer. We put together personalized certificates of completion for the high schoolers, we got some ice cream, and had a relaxing afternoon enjoying a picnic out on the oval.
Novel Selection Schema
Our interns stayed for 6 weeks, 7 weeks, and 11 weeks respectively. We were incredibly grateful for their choice to stay longer than required and to have their continued support with lab work and content creation for the website. By the end of the process they had become experts of PCR, gel extractions, mini-preps, cell-free protein expression, and much more. As they were able to gather expertise from working with a variety of lab members, by the end of the summer I personally would sometimes go to them for help with these protocols when I wanted tips and pointers.
Our survey data showed the following:

Additionally, our short answers sections showed the following:
Novel DE Application
Experiments
Our first experiment was to test AcrIIA4 against both dCas9 and dxCas9 in cell free. We devised the following test groups:

We used the PURExpress® In Vitro Protein Synthesis Kit from NEB and ran our test groups in duplicates for 5 hours. During this time, we monitored fluorescence every ten minutes in our plate reader with excitation set to 520 nm and emission measurement at 610 nm.


