GENETIC CIRCUIT
Our system is composed of 2 plasmids (pEHV01 y pCMR01) (Figure 1) , both plasmids co-transform using E. Coli as chasis creating a protein complex that serves as a "microbioreactor" which when in contact with vanilate activates, starting its transformation in cis, cis-muconic acid. The process uses bacterial microcompartments, which when confined a group of reactions potentially toxic in its interior, allows the bacteria to have a higher efficiency in this metabolic process.The process is regulated by pVanCC y p3B5C (promoters), which respond to initial phenolic compounds or derivatives of the metabolic route selected, this allows a regulation in phases which adapts to each enzyme specs.
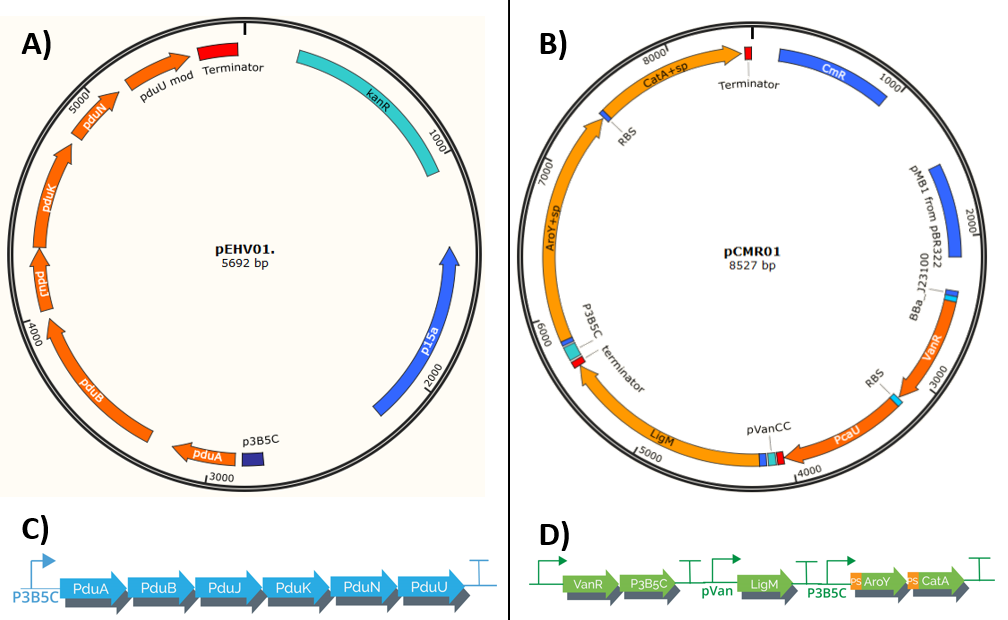
pEHV01 transports an operon which encodes a group of 6 proteins derived from S. enterica LT2 (<partinfo>K3317033</partinfo>) responsible for the formation of the Bacterial microcompartments (BMCs), structures formed by a thin protein shell (1). The final construct is under the regulation of the promoter p3B5C (<partinfo>K3317007</partinfo>), such promoter has operator sites to which the repressor protein pcaU (<partinfo>K3317005</partinfo>) binds, when interactings with protocatechuic acid (PCA) the protein allows the transcription (2). The final operon has a modification in the protein pduU(<partinfo>K3317014</partinfo>), the deletion of the first 17 amino acid residues leads to in an increase of the permeability of the Bacterial microcompartments without altering their stability.

The assembly of the plasmid pEHV01 (Figure 1A) began by cloning a fragment 543 bp (Figure 2) in a plasmid pSB3K3(<partinfo>K3317007</partinfo>), the fragment contains the promoter p3B5C a ribosome binding site (RBS) designed for the protein pduA. Inside the resulting plasmid an insertion can be performed of a group of 6 proteins (psuABJKNU) (Figure 3A) of S. enterica LT2 (NC_003197.1) of the plasmid pMCY30 provided by Ph.D Mimi C. Yung (Figure 3) through a cloning by homologous recombination as described by Jacobus et al 2015(3). The resulting plasmid (containing pSB3K3 promoter and psuABJKNU proteins) contains sites that can be amplified by PCR to perform a second homologous recombination (Figure 3B).
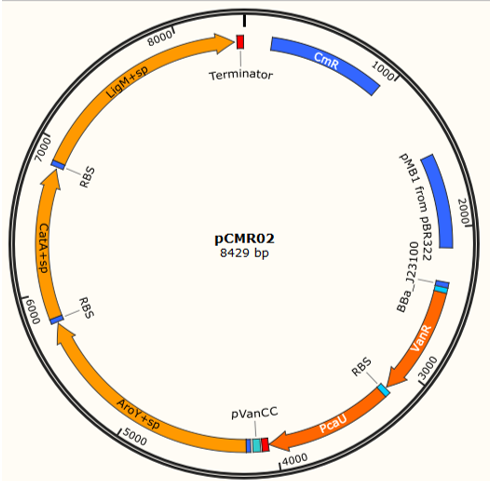
pCMR01 (Figure 1B) is a plasmid responsible for transport the metabolic pathway responsible of transform vanillic acid to muconic acid containing 3 enzymes: LigM , aroY y CatA. Furthermore, it contains the regulatory proteins VanR and pCau. The first enzyme, LigM , is regulated by the promoter PVanCC which allows its expression in the presence of vanilate which is the substrate of LigM transforming it in protocatechuic acid in the cytoplasm. The second and third enzyme which are regulated by a promoter sensitive to protocatechuic acid contain a signal peptide which is responsible for their internalization in the BMCs making the transformation of protocatechuic acid to catechol and afterwards to a cis, cis-muconic acid.
The assembly of the plasmid pCMR01 can be carried out through the method 3A Assembly, binding the generator of the regulatory proteins VanR and pCaU (<partinfo>K3317029</partinfo>) along with the operon responsible for the metabolic pathway (<partinfo>K3317030</partinfo>) within a vector pSB6c5.
A third plasmid named pCMR02 (Figure 4) was designed, through an enzymatic digestion in the sites Pvul of pCMR01 eliminating the protein LigM , its RBS, the corresponding terminator and the promoter pSB3K3. Performing a digestion in the sites HindeIII and XhoI the protein LigM carrying a signal peptide in its carboxil terminus which internalizes the enzyme in the BMCs can be inserted, leading to 3 enzymes ( LigM , aroY and CatA) (<partinfo>K3317032</partinfo>) regulated by the promoter pVanCC.
Bibliography
1:Jorda, J., Liu, Y., Bobik, T. A., & Yeates, T. O. (2015). Exploring bacterial organelle interactomes: a model of the protein-protein interaction network in the Pdu microcompartment. PLoS computational biology, 11(2), e1004067.
2:Meyer, A. J., Segall-Shapiro, T. H., Glassey, E., Zhang, J., & Voigt, C. A. (2019). Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nature chemical biology, 15(2), 196.
3:Jacobus, A. P., & Gross, J. (2015). Optimal cloning of PCR fragments by homologous recombination in Escherichia coli. PLoS One, 10(3), e0119221.