Contents
Design
Introduction and background
Bioeconomy transform lignocellulosic biomass into products with higher value, that may substitute the petroleum derivatives. To do this, first its necessary to separate lignin from cellulose with hidrolytic pre-treatments, with the objective of use glucose as a carbon source. However, lignin phase and sub products formed by this process has high toxic properties that complicate the biotransformations. Black liquor (BL) is the most concentrated phase, almost all by lignin derived components, called lignin derived inhibitory compounds (LDICs). The aromatic components in BL has a high energy, and the actual industrial use is burning it to produce heat and energy, which we may take advantage to produce bulk chemicals with higher value added products and environmental impact. To use BL as a substrate in an efficient way, we designed a metabolic network involving organelle engineering. To achieve this, we divided the project into three main parts:
- Selection of synthetic metabolic pathway. This pathway converts vanillic acid, a LDIC, into muconic acid, a nylon precursor.
- Organelle engineering. Protein engineering to encapsulate enzymes and enhance permeability of a bacterial microcompartment.
- Genetic circuit design. Would provide slightly regulation and optimization variables, as BMC assembly, growth and dynamic metabolic control.
Selection of the metabolic pathway.
To select the pathway, we established three objectives:
- The final product may be higher value added product that BL burning to produce energy.
- It must be valuable in the market
- The final product can also be extracted from petroleum derivatives.
BL composition analysis
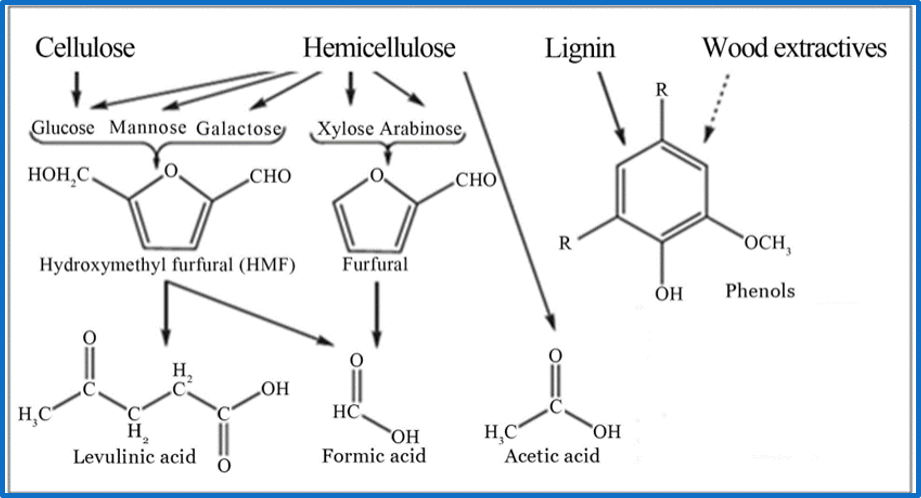
The LDICs are very diverse because their structure depends on the composition of lignin from de organic-vegetal source. In this case, we analyzed the source reported from kraft pulp, generated by the paper industry. The components that are present in higher quantities are guaiacol, catechol, vanillin, vanillic acid and isoeugenol. (1,2) Aromatic phenolic compounds are hydrophobically predominant. Most of them can cross lipid membranes easily, which causes their specific toxicity. Their IC50 varies from 0.2g/L until 2 g/L for vanillin and vanillic acid. These factors were important when designing the system. However, the selection of the first substrate would impact the overall system.
Synthetic metabolic pathways for revaluation purposes
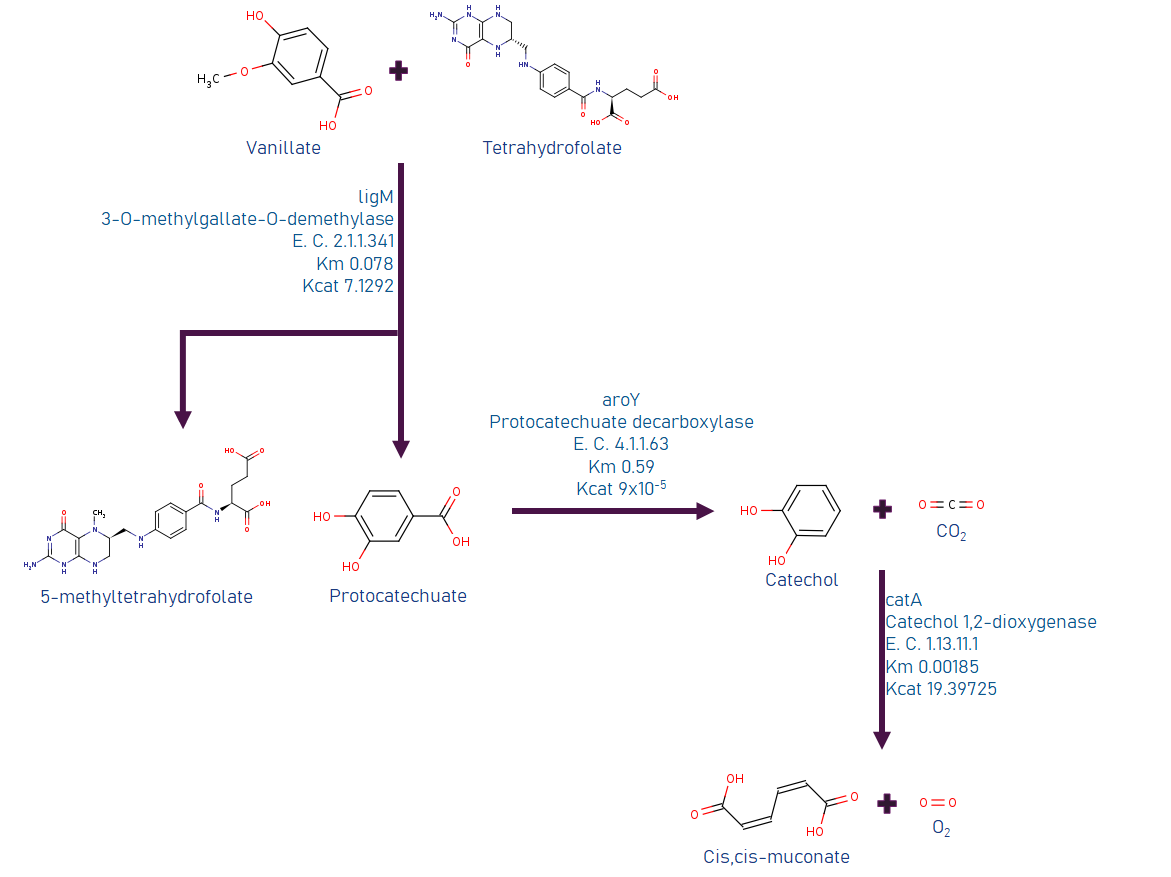
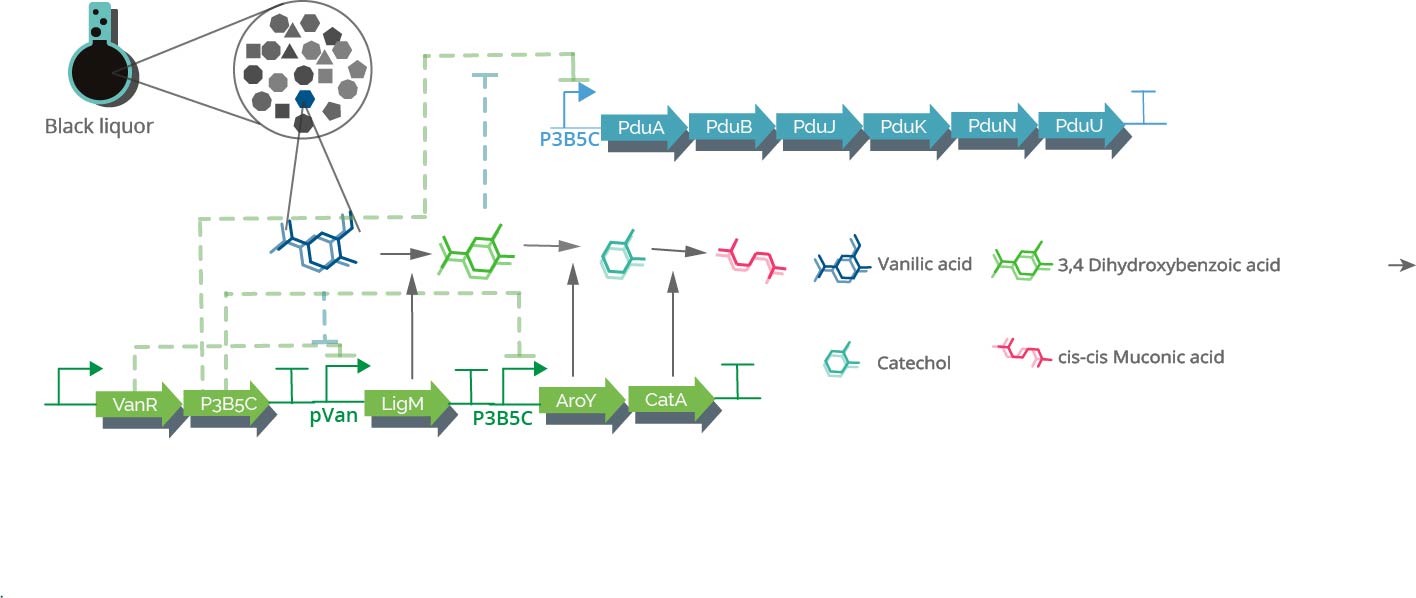
We investigated several ways to build or implement a synthetic route to use LDICs as a primary substrate, with the main goal of using them as raw material to synthesize other high-value products. As the LDICs are hard to work with; since there are not a lot of metabolic systems to implement new synthetic pathways. We first used software like RetroPath2.0 (3), a program that is useful to discover and predict new metabolic pathways; however, time and resources were limited. To understand and model metabolic systems, we eventually decided to select a constructed pathway reported before. We chose the synthetic pathway, which has a set of known enzymes and previously expressed in E. coli(4). The synthetic pathway uses as primary metabolite vanillic acid, as they oxidize to cis-cis muconic acid. Figure 2 (on the left side) shows information about the synthetic pathway. Cis-cis-muconic acid may be transformed in adipic acid, and caprolactam, which are used in nylon production. We design metabolic analysis experiments to detect metabolites and their intermediates in culture media. Using TLC as easy control and further quantitative data with HPLC-UV/Vis. (4),(5)
Protein engineering design of synthetic organelles
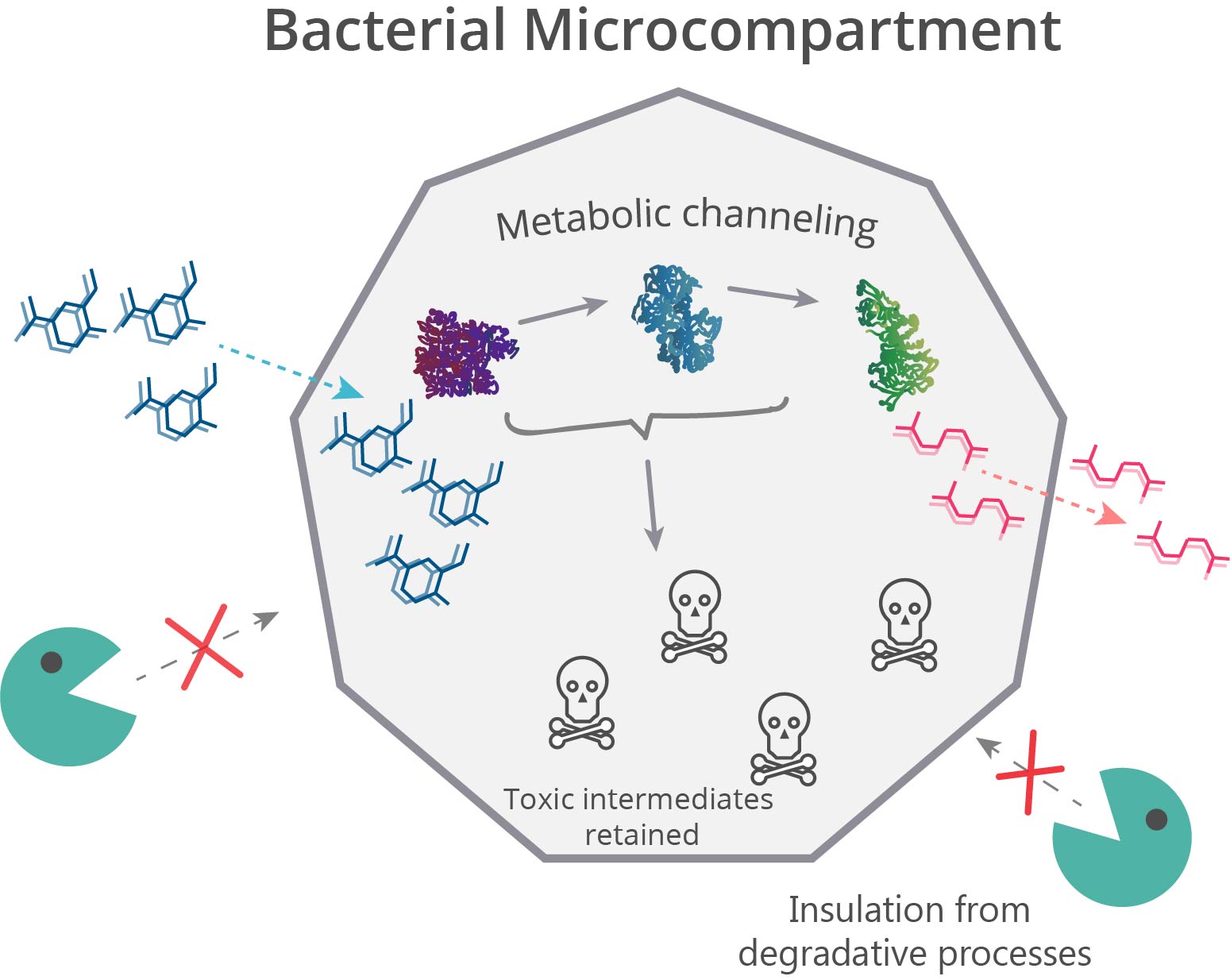
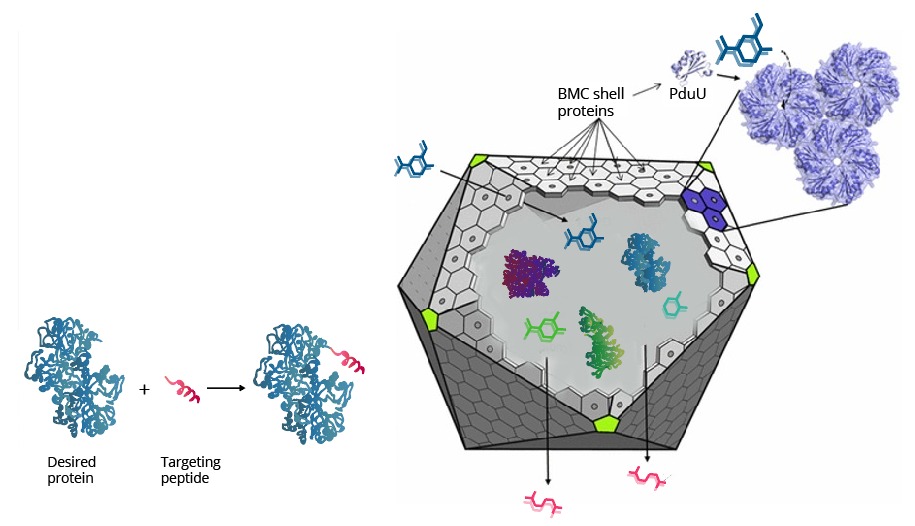
Spatial organization systems can develop a lot of advantages, one of them is compartmentalize chemical reactions, sequestering toxic intermediate and enhancing pathways flux (6) (Figure 3). As LDICs and the pathway selected has toxic intermediates, we implement the idea of encapsulation and isolation of metabolic systems using bacterial microcompartments (BMCs) (7–9). We explain first the basic functioning of BMCs, then which parts we design to use as a nanoreactor and organelle.
BMCs organelles, a useful and straightforward bioengineering chassis
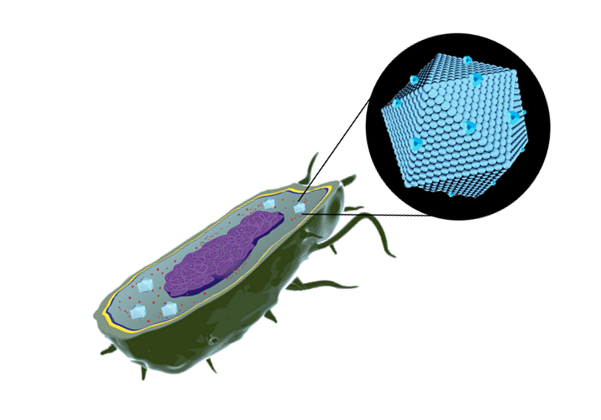
BMCs are protein-based organelles. They are composed of multiple protein subunits, which self-assemble into an icosahedron complex between 100-200 nm of diameter. This structure can be compared to some viral capsids, and they can direct proteins inside through an encapsulation-tag peptide. When the proteins encapsulate enzymes and pathways, they are called metabolosomes, and different subunits permits the metabolite diffusion through a pore . Their genomic organization is an operon of several genes and can be used to make easy organelle engineering, compared to lipid-based ones (10,11).
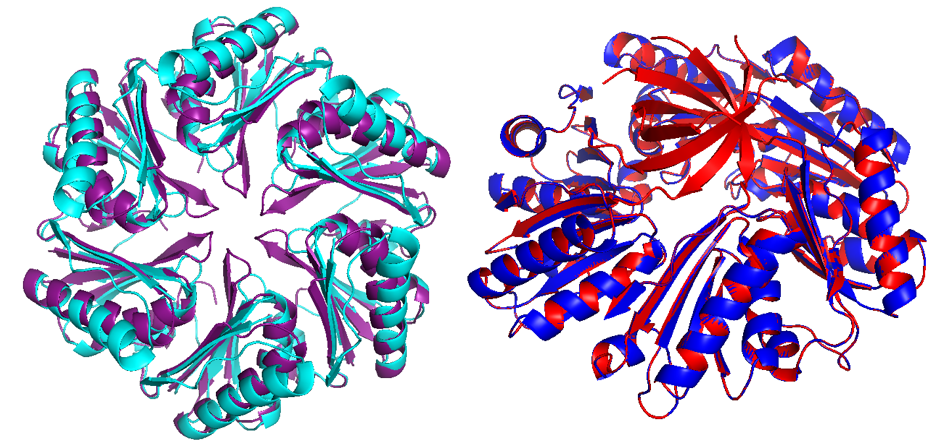
Different compartmentalization systems have been described in the past. We could have used Propanediol Utilization System (Pdu), ethanolamine utilization (Eut) and carboxysome (Ccm). We chose Pdu form Salmonella enterica, which is the best described in the literature.
Function in nature
On the right side, the representation of Pdu system from Salmonella enterica. The shell is conformed by Pdu units which most are self-assembly protein hexamers (represented as PduA), and they have a pore with selectivity for substrates. Wild type Pdu has selectivity to propanediol, and inside the shell, it encapsulates enzymes and cofactors necessary to produce 1-propanol, an energetic pathway. Protein encapsulation is possible by an encapsulation peptide (EP) that redirects protein inside(Left side). We use these principles to create a new metabolosome.
Subunits
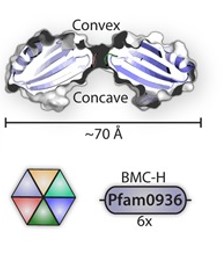
Pdu has several different units with the same structural and predominant domain called Pfam0936 (9) (Figure 7) to form empty shell complex, characteristic of BMCs systems. Pfam0936 permits interaction with other subunits in a zipper-like interaction and its highly stable. All units have convex and concave faces, corresponding with outer and inner shell faces respectively. This particularity has an impact on pore selectivity and shell assembly stability. In our system, there are only seven necessary units: PduA, PduB, PduU, PduJ, PduK, and PduN… pduABJKNU (14). We asked for an operon constructed by Mimi, and collaborators (23).
Shell proteins can be classified into different types as the roles in BMCs. BMC-H, BMC-P depending on their structure(11). BMC-H: PduA and PduJ, a hexameric protein which can form pores, PduU is a permuted pore with structural co-localization function in the cytoskeleton. BMC-T: PduB, a trimeric protein with pseudohexameric symmetry, which can form pores, and BMC-P: PduN, a pentameric protein with particular structural function.
Structural analysis for viable pore and protein engineering
BMC permeability engineering is one of the most challenging tasks actually in synthetic BMCs as the pore selectivity has been determined by two essential factors, size and electrostatic surface interaction with metabolites(10,15). To determine the permeability of our desired metabolites we need to analyze different pores.
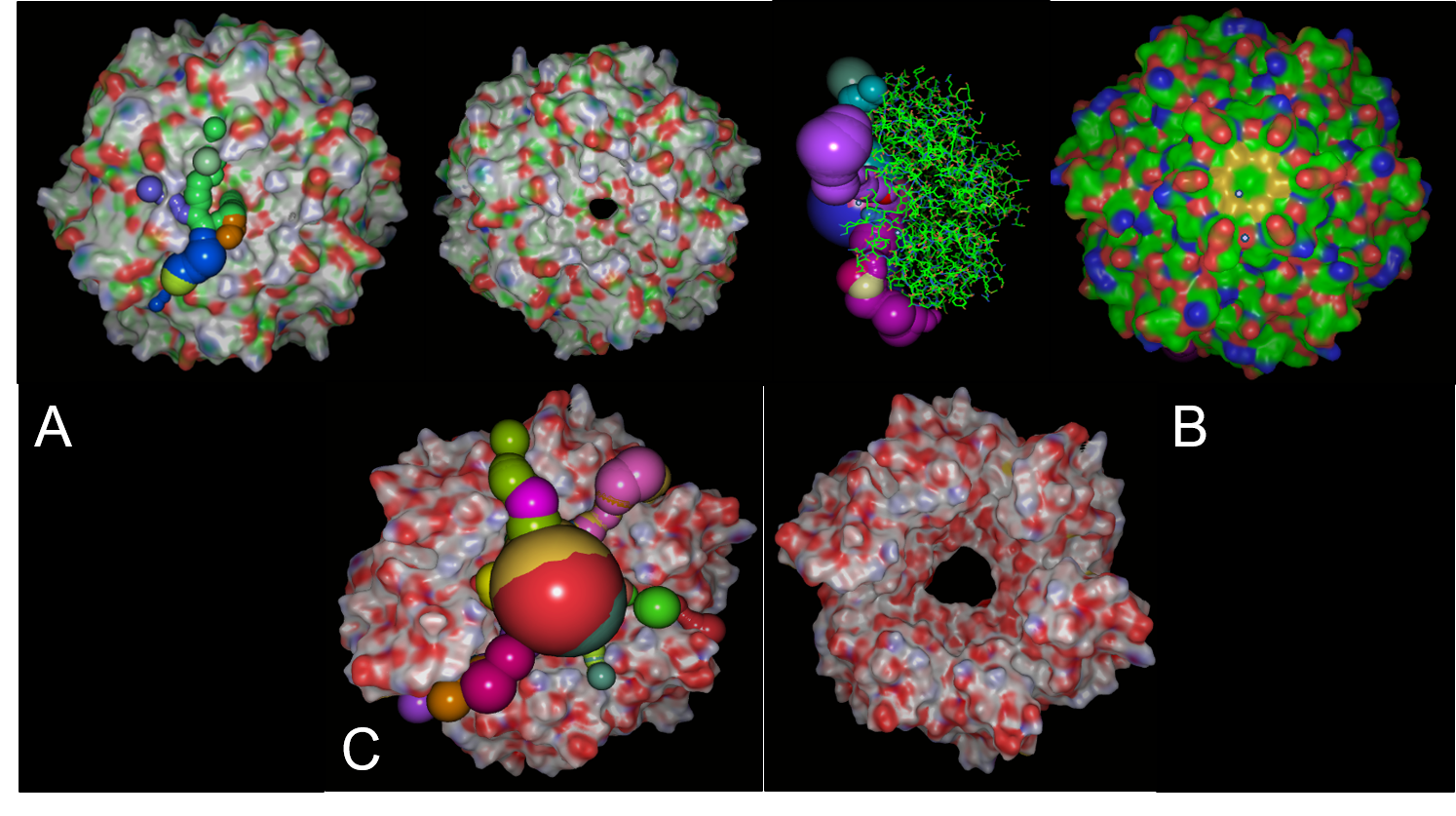
PduA, PduB and PduJ (PduABJ) are the most abundant shell protein, with similar function and structure. They had a central pore with similar width that permits the pass of metabolites through them, which naturally is propanediol. However, these are the most prominent pores in BMCs systems reported. Indeed, this characteristic may not be a deterministic way of knowing if it is porous with the width enough for vanillate, catechol, protocatechuate and other compounds pass trough. We use CaverSuite(16–18) and Molecular Dynamics Simulations in GROMACS software to determine these parameters important in BMC engineering.
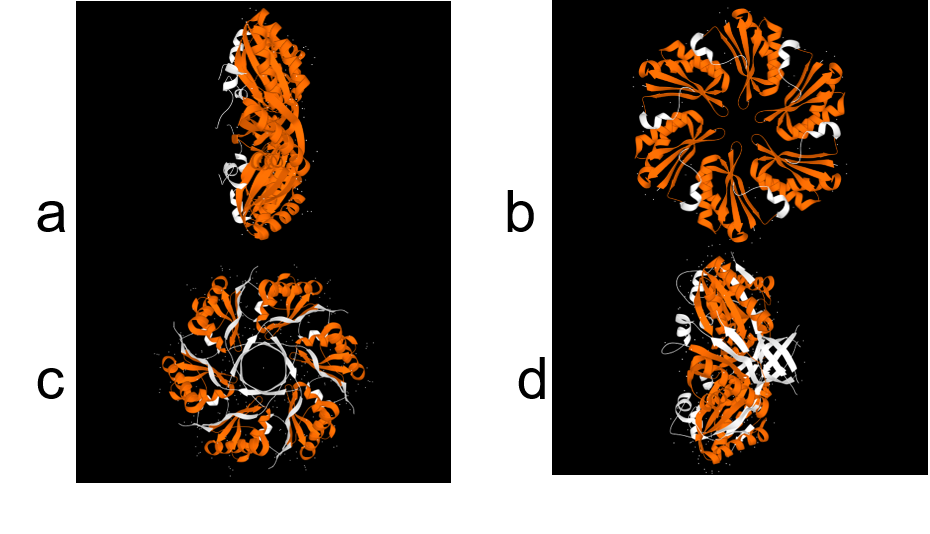
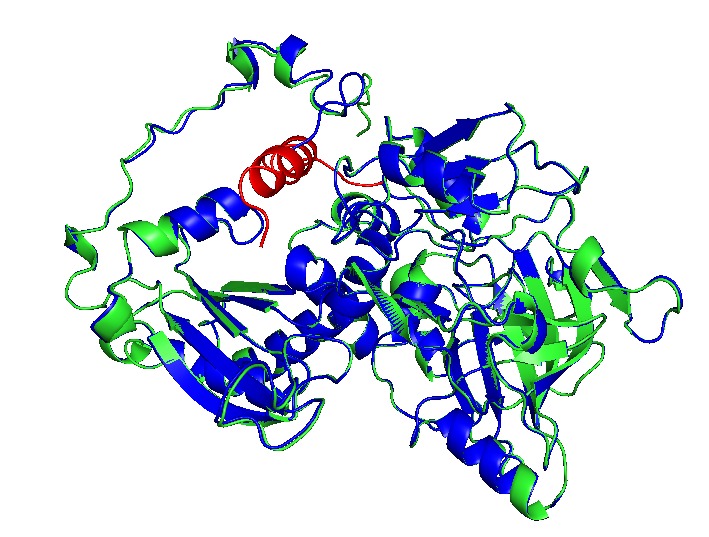
In structural analysis, it is reported that for all shell monomer-monomer and subunit-subunit interactions, is essential the Pfam0936 domain (represented in orange, figure 8). An α Helix (denoted in white in the concave face, figure 8) are motifs with the interaction with the encapsulation peptide. In PduABJ the subunits are almost fully composed by this domain. In the structural view of other subunits, we found particularities in PduU subunit, one of them is that it has an additional domain (not officially reported as a Pfam) and distinct folding which makes possibly enhance permeability. PduU has a β barrel (denoted in white in the convex face in PduU, figure 8) which has been computational determined that interact with PduV, at the same time this protein complex associates with the cytoskeleton to attach in a specific place in the cytoplasm. However, the shell can assembly without PduV, corresponding to the β barrel of PduU do not have an active function in the shell formation. (19)
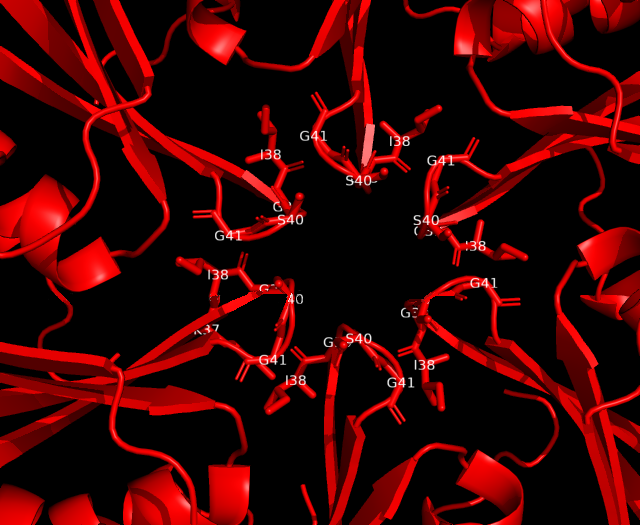
Analysis of cavity, electrostatic surfaces and tunnel generation with possible trajectories were made with CaverAnalyst software, indicating the real pore size in PduA are approximately 2.8 Å, almost the size of their selective metabolite propanediol. Residues T38, G39, S40, G41 that conform a loop in the centre pore, provide all their particular characteristics to the pore. Polar groups and positively charged pore have an affinity to negatively-charged molecules, like propanediol.
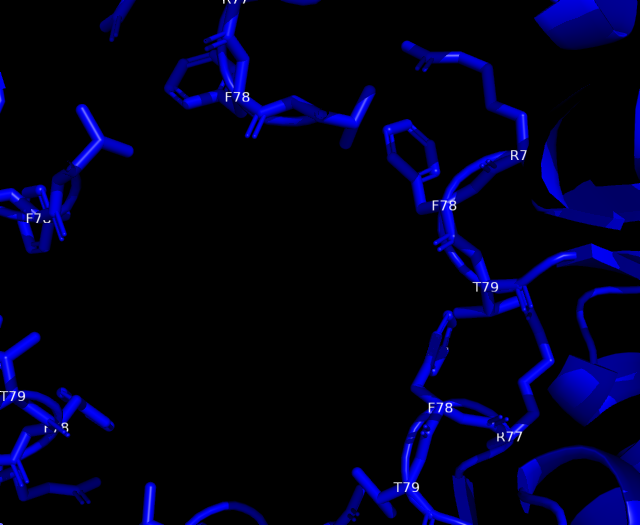
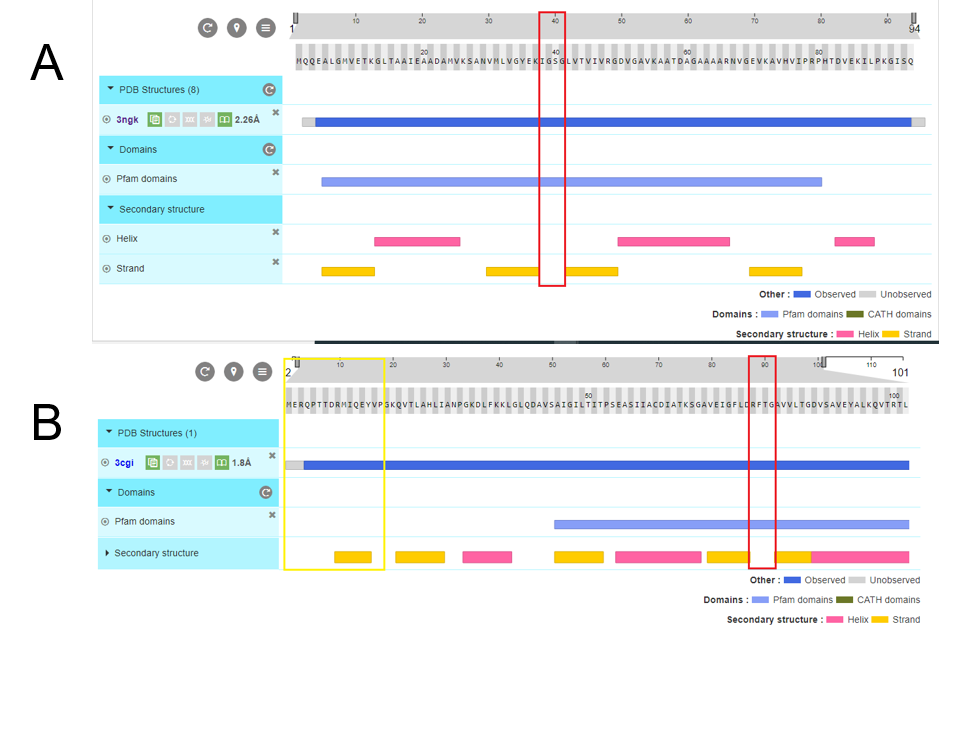
Fig 10.Structure analysis of A. PduA and B. PduU. Showed sequence, secondary structures, and pfam domains (the only one is pfam 0936 denoted in the light blue bar). Red rectangles marked the pore residues in similar ubication of the domain, however there are structural differences in residues corresponding to the pore. PduU shows a larger sequence and the yellow rectangle is denoted the β barrel, in N terminal, of 15 residues long.
The sequence showed that almost all the protein is formed by their main domain, pfam0936, which is challenging to make a rational protein engineering to enhance the pore permeability, because mutations in secondary structures may make an unstable unit and unfolding of the protein. Free energy analysis of protein allows only loops to mutate them without unstabilize the protein, but it would-be high-risk mutations. However, PduU subunit its a candidate to engineering permeability, because it has the biggest pore, and it is possible the engineering in β barrel that permutes the pore.
Modification of PduU subunit to engineer pore permeability of Pdu
We made deletion of 15 residues in the N-terminal sequence of PduU, creating PduUmd variant. β barrel was deleted, and the pore with bigger size and nonpolar partially charged pore may augment BMC permeability.
Fig. 12 Structural differences in PduA/PduUmd (Left) and PduU/Pdumd. Light blue PduUmd and violet PduA. Modelling demonstrates wide pores in the loops of the centre pore; however, their structures do not align accurately. (Right) Alignment between PduU (red) and PduUmd (blue), the pore is permuted by the β barrel which does not interact with pfam0936 domain.
We use phyre2 and I-TASSER software to predict tertiary structures, from the deleted sequence. However, has the biggest deletion do not clarifies the stability of the protein. We proceed to make stability analysis by computational methods described in simulations-modeling page, as a part of design process. In resume, we use molecular dynamics simulations and docking softare, the analysis confirms that the complex can be formed and being functional.
Fig.13 It is showed 3 proteins, after their solvation (except for PduU) in GROMACS, which represent a realistic view of the pore. On the left of every set of images are results of CaverAnalyst, which are tunnel surfaces. Every possible tunnel is showed in different colors and get joined in the pore. On the right side, the surface model of every protein. A. PduA B. PduU C PduUmd.As its showed, PduUmd gets a bigger pore, compared to PduA and it is not permuted like PduU wild type unit.
We can use experimental wet lab work to demonstrate the different units assembled. There are BMC extraction protocols with easy steps, further we can use SDS-PAGE and Atomic Force Microscopy to analyze the complex formed, encapsulated enzymes and subunits present. Permeability differences can be measured by titers in culture media of muconic acid and vanillic acid, also proven toxicity assays. We expect higher titers than reported, by the flux sequestration and toxic diminishing, however, energy demand of expression systems may slow bacterial metabolism. (8),(20)
Encapsulation peptides
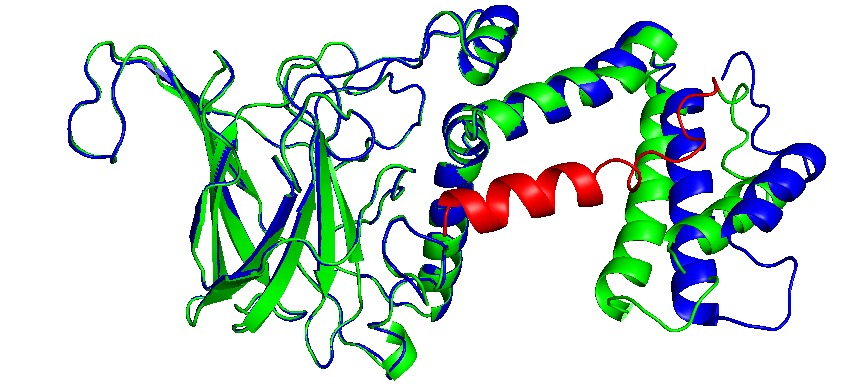
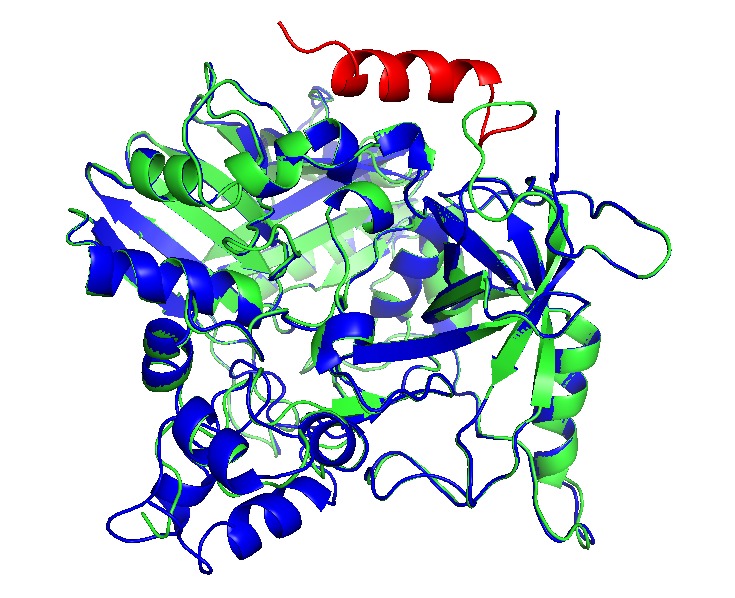
Proteins can be encapsulated by signal peptide, named as Encapsulation Peptide (EP).(14,21) EPs may vary between wild type proteins as PduD, PduP and other enzimes, which is 18 to 20 residues in alpha helix conformation. It is highly conserved and may interact with several units in the concave face of Pdu shell units, directing a protein aggregate inside the BMC. (22)Also, these EP can interact with other EPs, augmenting the encapsulation rate (8,20). The interactions with N terminal of PduP and C-terminal of PduA was determined, indeed, we extract EP from PduP with a 19 long residues. The linker sequence may be required and important in the attachment of new proteins. then a short linker G4S was added.
Enzymes with EP
We add the selected EP (See add sequence structures) to the N- teminal and C- terminal sequences, depending their structural conformation using Phyre2 to modelling and predict tridimensional structures. CatA and AroY seems that N-terminal added EP (name) do not interact with catalityc sites, as LigM the signal peptide interact between monomers and get “buried” between them, blocking catalytic sites. EP was then added in C-terminal in a way to prevent this conformation.
Showed enzymes in alignment, original in blue, with EP in green and the EP is in red.
Assembly
BMCs have the property of self-assembling. Analysis of the residues in the interaction zone between units has a “zipper” mechanism involving polar interactions(12). This residues are highly conserved, which makes possible hybrid BMCs. However, there are several variables involving the right assembling of BMCs, some are the protein-encapsulated concentration and BMCs subunits ratio. As the assembly depends in equilibrium between EP and subunits interactions, the size and stability depend in the expression rate of proteins. This is a problem afforded in the construction of genetic circuit in which is the regulation of expression can be slightly controlled. However, attempts reported in encapsulated protein vary between 1-5% of total protein expressed.(23) Our circuit constructed may be a great way to optimize the encapsulation and BMC stability, preventing formation of other types of structures, like nanotubes or inclusion bodies.(24)
Expession and regulation systems: Genetic circuits
Importance of expression regulation in our system:
- Assembly and encapsulation control(25)
- Dynamic control of metabolic systems(26)
- Strategies for metabolic optimization(27)
We decide to use 2 expression systems in 2 plasmids, as a modular BioBrick for BMC expression and pathway expression. With this approach, anyone can use our BioBricks as nanoreactor, and design a pathway inside with EP. However, some aspects are of exclusive importance, even for the study of BMC nature, like the right equilibrium of enzymes and Pdu subunits, EP dynamics and metabolic control.
Promoters: Selection and design
We decide to use 2 expression systems in 2 plasmids, as a modular BioBrick for BMC expression and pathway expression. With this approach, anyone can use our BioBricks as nanoreactor, and design a pathway inside with EP. However, some aspects are of exclusive importance, even for the study of BMC nature, like the right equilibrium of enzymes and Pdu subunits, EP dynamics and metabolic control.
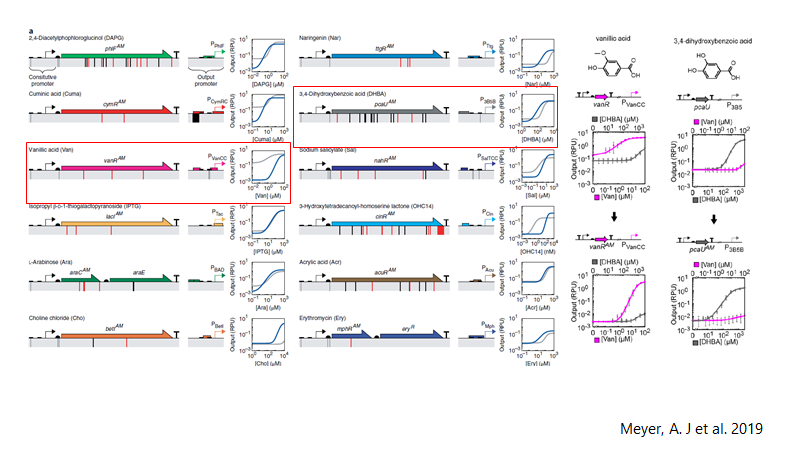
Promoters reported and optimized by Meyer, et al. (28) was selected PVanCC and P3B5C ,which are positively regulated by vanillic acid and protocatehuate (2,3 dihidroxybenzoic acid, DHBA) acting as inducers. These promoters are regulated by VanR and PcaU repressor proteins respectively (PVanCC-Vanillate and P3B5C-DHBA). If we remember the pathway, those are two pathway intermediates, that each one will induce a module for synthetic pathway and BMC system, which allows to control expression of enzymes and BMC in a harmonic way. This may completely design and modeled in a math model of expression, as the promoters are characterized in Relative Promoter Units (RPU), we have accurate quantitative data of them. Also, the promoters are optimized to diminishing crossing over signaling between them and leakiness.
Promoters reported by Meyer, et al (28)Vanillate is present in black liquor, so it must activate the system with the presence of these small molecules. As the vanillate induces the expression of Vanillate-O-demethylase, it will demethylated by the enzyme, producing DHBA, which actives the second module of expression, that is Protocatehuate Decarboxylase, and Catechol 1,2 dioxygenase, and Pdu subunit cassette. Vanillate is also cheaper that most of inducers.
Ribosome binding sites, RBS calculator and standardized promoters as a predictive biosystems engineering.
RBS is an important part of protein expression systems. As we look into a more accurate expression systems, we use a useful tool to predict quantitative data from RBS sequences, the RBS calculator.(28–30) RBS from registry parts are in relative units of biobricks sequences. However, those sequences are only a small part of all the complex implications for the translation initiation, in which coding sequence has a definitive impact on translation rate, as the mRNA foldings can modulate the ribosome attachment. For this reason, a RBS cannot being standardized accurately. We work in other possibility system for characterizing or reporting the data in the best, useful and accurate way, into proposal for a model. However, we registered coding sequences with RBS calculator.
Overview of genetic circuit.
Construction
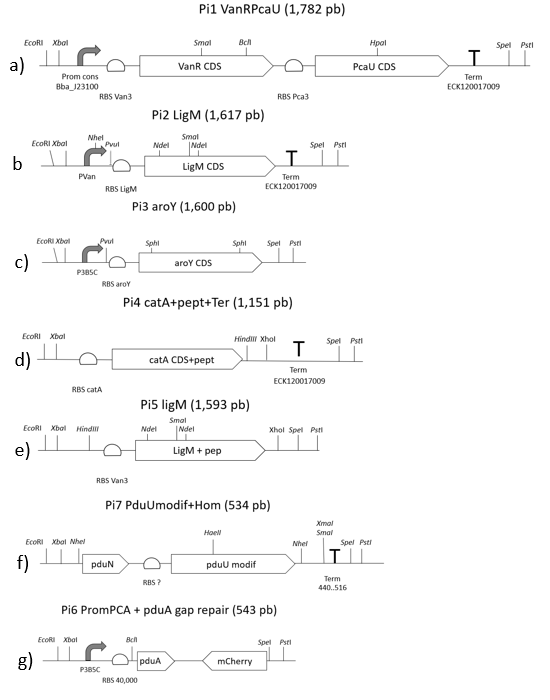
For the assembly of both plasmids that make up our system (pEHV01 and pCMR01) we start from the design and synthesis of 7 double-stranded linear fragments not bigger than 2000 bp provided by IDT (Figure 11E), these fragments have restriction sites in its ends own prefix and suffix of biobricks.
Mentioned fragments (Figure 11) were destined to cloning in vector pSB1C3 provided by iGEM. The linear fragments 1, 2, 3, and 4 correspond to the metabolic pathway pieces (<partinfo>K3317030</partinfo> <partinfo>K3317029</partinfo>), these fragments are the ones responsible for creating the metabolic route and regulation of our own system. (Figure 12B)
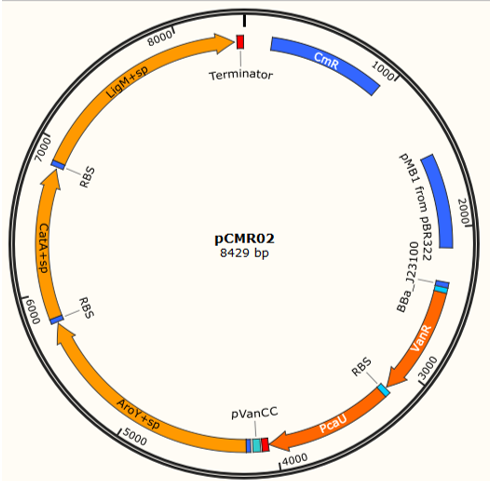
We used 3A assembly method to built fragment to the pieces (<partinfo>K3317030</partinfo> <partinfo>K3317029</partinfo>) in a vector with a multicopy origin pSB1C. Once the fragment is cloned, in must being transfer to a vector pSB6C5, forming our final plasmid pCMR01 (Figure 12B). The pSB6C5 skeleton is used given by his replication origin which is composed of medium copies,in addition to the presence of XhoI restriction sites. The pSB1C3 skeleton prevents its subsequent modification to form the alternative plasmid pCMR02 (Figure 13)
Linear fragments 6 and 7 correspond to the assembly of plasmid pEHV01 (Figure 12A). The <partinfo>K3317034</partinfo>, piece responsible generating the bacterial microcompartment in our system. Fragments 5 and 7 are designed to perform a cloning by homologous recombination (31), this is because BMC-forming protein operon from plasmid pMCY30 has sites that make it impossible to assemble by 3A assembly (23). The final vector carries the piece <partinfo>K3317034</partinfo> is pSB3K3, this to allow its compatibility with pSB6C5 when carrying a different selection marker and replication sources that belong to different groups.
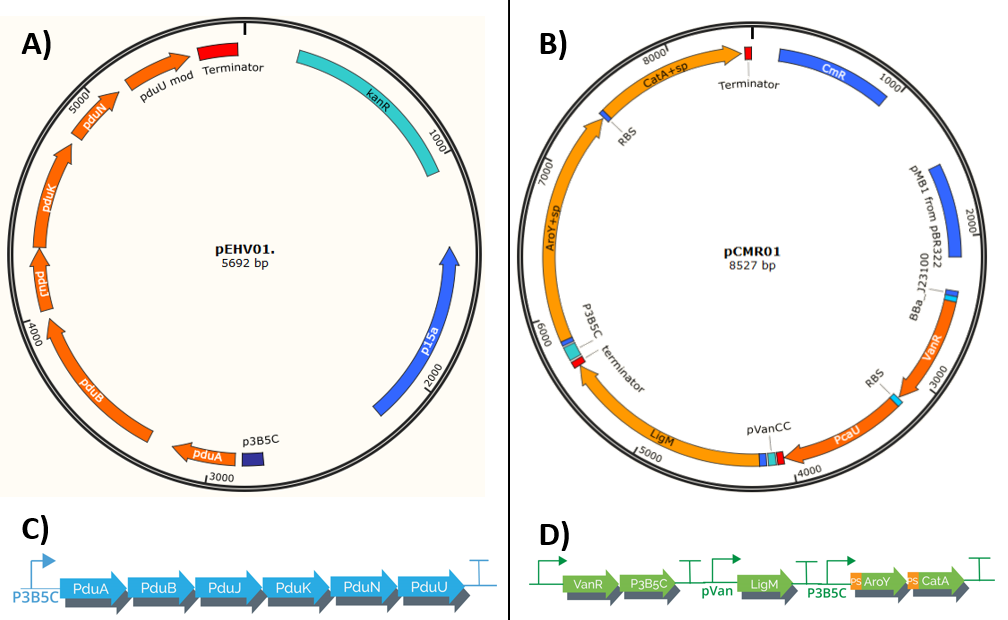
Linear fragment 5 (Figure 11E) is used to build an alternate plasmid, pCMR02 (Figure 13) carrying the part (<partinfo>K3317032</partinfo>) which has the 3 enzymes of our metabolic route modified with the signal peptide that introduces them to BMC. The generation of plasmid pCMR02 is from modifications in plasmid pCMR01 which has two PvuI cutting sites, the first one is located immediately below the pVanCC promoter (Figure 11B) and the second downstream of the p5B3C promoter (Figure 11C), in this way, we substitute the sequences corresponding to the ribosome binding site of the LigM protein, the LigM protein itself and a terminator are eliminated. Also, plasmid pCMR01 has HindCIII and XhoI restriction sites downstream of the coding sequence for the modified catA protein (Figure 11D) in which it inserts the linear piece 5.
Construction Resume
2 pathway routes. pCMR02: This plasmid is constructed with the objective of measure the titers of encapsulation of the 3 enzymes under control of Vanillate and Protocatehuate, as LigM uses Tetrahydrofolate as a substrate, which may not pass through the pore. pCMR01: This plasmid is constructed with the objective of mantain LigM outside the BMC system, with CatA and AroY encapsulated, this is because LigM uses tetrahydrofolate as a substrate, which may not pass through the pore. We can measure the predictions used in model and previous design sections, and optimize the metabolic systems depending in substrate permeability.
Pdu systems: pEHV01: Engineered Pdu operon.
References
1. Milan D, Peal D. (12) Patent Application Publication (10) Pub . No .: US 2002/0187020 A1. Vol. 1. 2013.
2. Ponnusamy VK, Nguyen DD, Dharmaraja J, Shobana S, Banu JR, Saratale RG, et al. A review on lignin structure, pretreatments, fermentation reactions and biorefinery potential. Bioresour Technol [Internet]. 2019;271(August 2018):462–72. Available from: https://doi.org/10.1016/j.biortech.2018.09.070
3. Delépine B, Duigou T, Carbonell P, Faulon JL. RetroPath2.0: A retrosynthesis workflow for metabolic engineers. Metab Eng. 2018;45(December 2017):158–70.
4. Wu W, Dutta T, Varman AM, Eudes A, Manalansan B, Loqué D, et al. Lignin Valorization: Two Hybrid Biochemical Routes for the Conversion of Polymeric Lignin into Value-added Chemicals. Sci Rep. 2017;7(1):1–13.
5. Sutherland JB, Crawford DL, Pometto AL. Catabolism of substituted benzoic acids by Streptomyces species. Appl Environ Microbiol. 1981;41(2):442–8.
6. Castellana M, Wilson MZ, Xu Y, Joshi P, Cristea IM, Rabinowitz JD, et al. Enzyme clustering accelerates processing of intermediates through metabolic channeling. Nat Biotechnol [Internet]. 2014;32(10):1011–8. Available from: http://dx.doi.org/10.1038/nbt.3018
7. Giessen TW, Silver PA. Encapsulation as a Strategy for the Design of Biological Compartmentalization. J Mol Biol [Internet]. 2016;428(5):916–27. Available from: http://dx.doi.org/10.1016/j.jmb.2015.09.009
8. Huber I, Palmer DJ, Ludwig KN, Brown IR, Warren MJ, Frunzke J. Construction of Recombinant Pdu Metabolosome Shells for Small Molecule Production in Corynebacterium glutamicum. ACS Synth Biol. 2017;6(11):2145–56.
9. Plegaria JS, Kerfeld CA. Engineering nanoreactors using bacterial microcompartment architectures. Curr Opin Biotechnol [Internet]. 2018;51:1–7. Available from: http://dx.doi.org/10.1016/j.copbio.2017.09.005
10. Cai F, Sutter M, Bernstein SL, Kinney JN, Kerfeld CA. Engineering bacterial microcompartment shells: Chimeric shell proteins and chimeric carboxysome shells. ACS Synth Biol. 2015;4(4):444–53.
11. Lee MJ, Palmer DJ, Warren MJ. Biotechnological Advances in Bacterial Microcompartment Technology. Trends Biotechnol [Internet]. 2019;37(3):325–36. Available from: http://dx.doi.org/10.1016/j.tibtech.2018.08.006
12. Kerfeld CA. A bioarchitectonic approach to the modular engineering of metabolism. Philos Trans R Soc B Biol Sci. 2017;372(1730).
13. Slininger Lee MF, Jakobson CM, Tullman-Ercek D. Evidence for Improved Encapsulated Pathway Behavior in a Bacterial Microcompartment through Shell Protein Engineering. ACS Synth Biol. 2017;6(10):1880–91.
14. Sargent F, Davidson FA, Kelly CL, Binny R, Christodoulides N, Gibson D, et al. A synthetic system for expression of components of a bacterial microcompartment. Microbiol (United Kingdom). 2013;159(PART11):2427–36.
15. Park J, Chun S, Bobik TA, Houk KN, Yeates TO. Molecular Dynamics Simulations of Selective Metabolite Transport across the Propanediol Bacterial Microcompartment Shell. J Phys Chem B. 2017;121(34):8149–54.
16. Daniel L, Marques SM, Kozlikova B, Jurcik A, Bednar D, Stourac J, et al. CAVER Analyst 2.0: analysis and visualization of channels and tunnels in protein structures and molecular dynamics trajectories. Bioinformatics. 2018;34(20):3586–8.
17. Filipovič J, Vávra O, Plhák J, Bednář D, Marques SM, Brezovský J, et al. CaverDock: A Novel Method for the Fast Analysis of Ligand Transport. 2018;1–24. Available from: http://arxiv.org/abs/1809.03453
18. Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, et al. CAVER 3.0: A Tool for the Analysis of Transport Pathways in Dynamic Protein Structures. PLoS Comput Biol. 2012;8(10):23–30.
19. Jorda J, Liu Y, Bobik TA, Yeates TO. Exploring Bacterial Organelle Interactomes: A Model of the Protein-Protein Interaction Network in the Pdu Microcompartment. PLoS Comput Biol. 2015;11(2):1–23.
20. Nichols TM, Kennedy NW, Tullman-Ercek D. Cargo encapsulation in bacterial microcompartments: Methods and analysis [Internet]. 1st ed. Vol. 617, Methods in Enzymology. Elsevier Inc.; 2019. 155–186 p. Available from: http://dx.doi.org/10.1016/bs.mie.2018.12.009
21. Lawrence AD, Frank S, Newnham S, Lee MJ, Brown IR, Xue WF, et al. Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth Biol. 2014;3(7):454–65.
22. Lee MS, Tullman-Ercek D. Practical considerations for the encapsulation of multi-enzyme cargos within the bacterial microcompartment for metabolic engineering. Curr Opin Syst Biol [Internet]. 2017;5:16–22. Available from: https://doi.org/10.1016/j.coisb.2017.05.017
23. Yung MC, Bourguet FA, Carpenter TS, Coleman MA. Re-directing bacterial microcompartment systems to enhance recombinant expression of lysis protein E from bacteriophage φX174 in Escherichia coli. Microb Cell Fact. 2017;16(1):1–17.
24. Hagen AR, Plegaria JS, Sloan N, Ferlez B, Aussignargues C, Burton R, et al. In Vitro Assembly of Diverse Bacterial Microcompartment Shell Architectures. Nano Lett. 2018;18(11):7030–7.
25. Faulkner M, Zhao LS, Barrett S, Liu LN. Self-Assembly Stability and Variability of Bacterial Microcompartment Shell Proteins in Response to the Environmental Change. Nanoscale Res Lett. 2019;14:0–7.
26. McNerney MP, Watstein DM, Styczynski MP. Precision metabolic engineering: The design of responsive, selective, and controllable metabolic systems. Metab Eng. 2015;31:123–31.
27. Sweetlove LJ, Fernie AR. The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat Commun [Internet]. 2018;9(1). Available from: http://dx.doi.org/10.1038/s41467-018-04543-8
28. Meyer AJ, Segall-Shapiro TH, Glassey E, Zhang J, Voigt CA. Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nat Chem Biol. 2019;15(2):196–204.
29. Espah Borujeni A, Channarasappa AS, Salis HM. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res [Internet]. 2013 Nov 14;42(4):2646–59. Available from: https://doi.org/10.1093/nar/gkt1139
30. Borujeni AE, Cetnar D, Farasat I, Smith A, Lundgren N, Salis HM. Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences. Nucleic Acids Res. 2017;45(9):5437–48.
31. Salis HM. The ribosome binding site calculator. Methods Enzymol. 2011;498:19–42.