Contents
Modeling and Molecular Dynamics
Molecular Dynamics (MD) is a computational (numerical) method of simulation, in which the equations of motion are solved numerically to study the physical movement of atoms and molecules in a system. There are several programs that can perform Molecular Dynamics (MD) Simulations: MacroModel, LAMMPS, NAMD, etc. are some examples, but in this work GROMACS (gmx), a high performance MD program, was used to solve the equations of motion and Visual Molecular Dynamics (VMD) was used to construct the systems.
Modeling
As explained in Design, protein engineering was needed to modify one of the proteins of the Bacterial Microcompartment (BMC) to increase the permeability of substrate and enable it to pass through the proteinic shell. Specifically, an amino-acidic deletion in the n-terminal of the pduU protein (one of the structural proteins of pdu BMC) was desired.
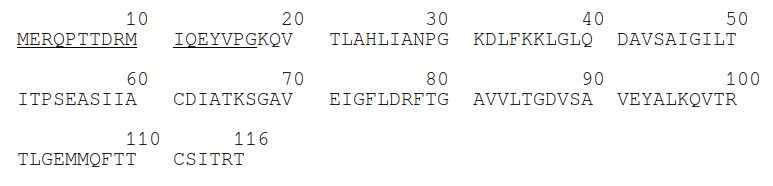
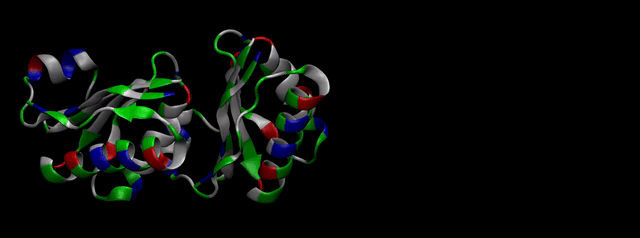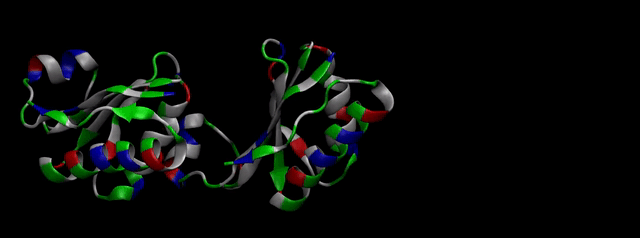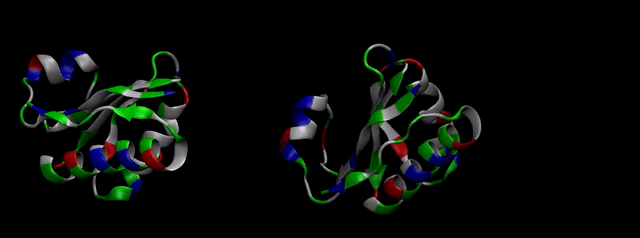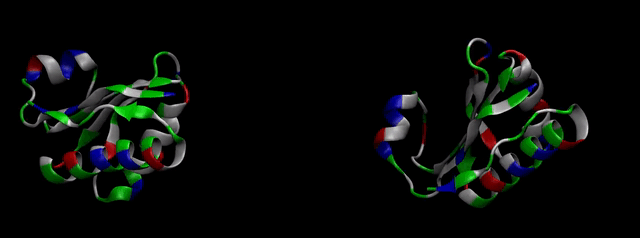
So, we took the FASTA format of the protein (Figure 2)...
... and deleted the first 17 amino-acids. We chose to delete those specifically because that modification had already been reported in an article by (J. Jorda 'et. al.' 2015)for other purposes.
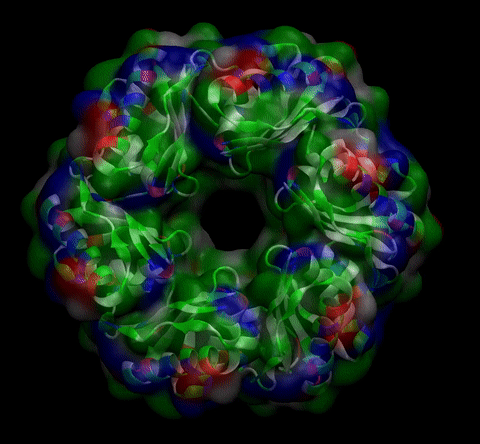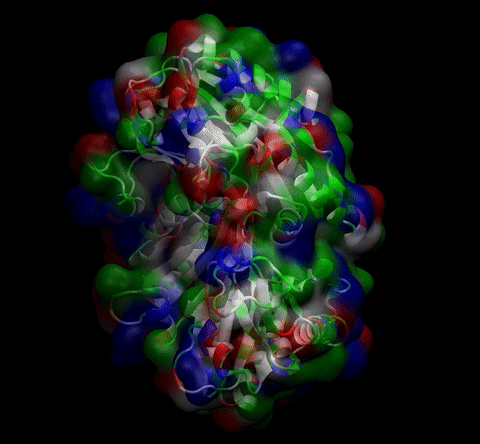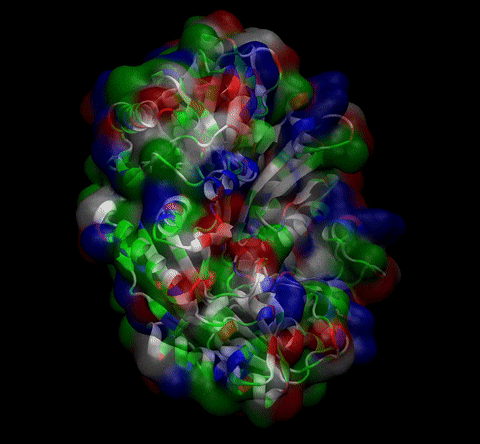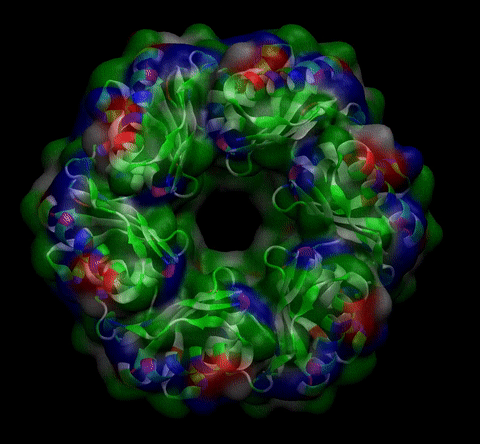
We then proceeded to evaluate it’s tertiary conformation with SwissModel, which uses homollogy modeling to predict the tertiary conformation of amino acidic sequences, and we obtained the predicted 3D conformation (1). It's easy to see that the porous is bigger now:). And thats VERY cool. Very nice.
MD Simulatoins
MD simulations are divided in 2 parts:
- Stability of modified protein verification
- Permeability of new porous
Stability
We estimated that this 3D array would be stable because, as explained in Design, most of that remaining domain is a very conserved one, but we still wanted to prove it through Molecular Dynamics simulations, which we performed using GROMACS package (Bekker 'et. al' 1993) through
2 methods:
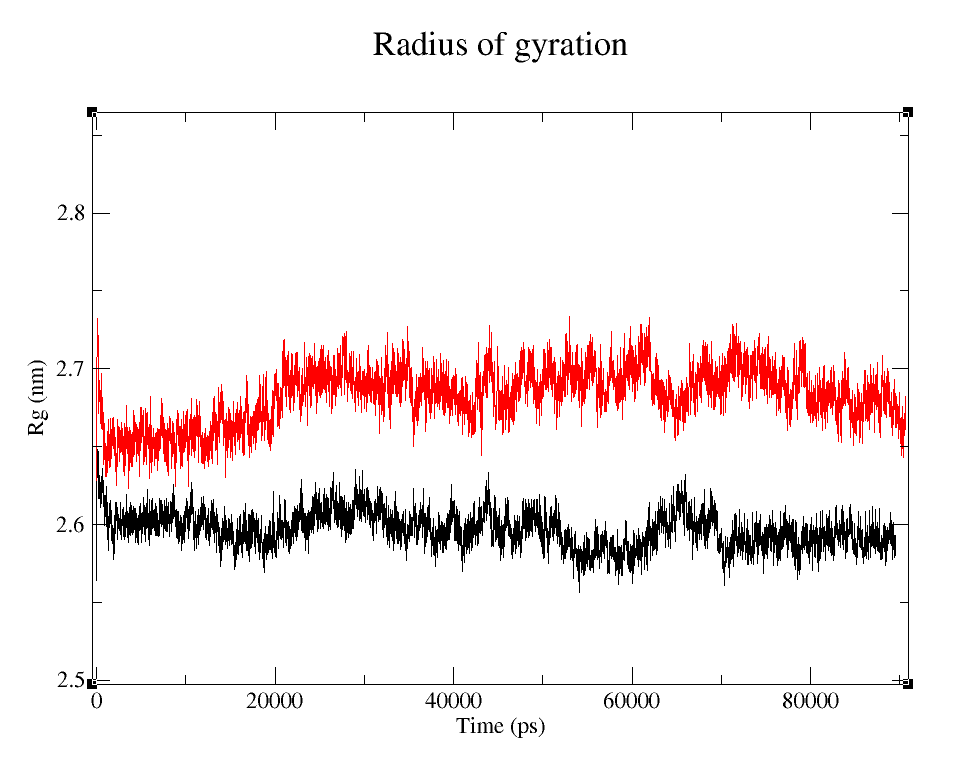
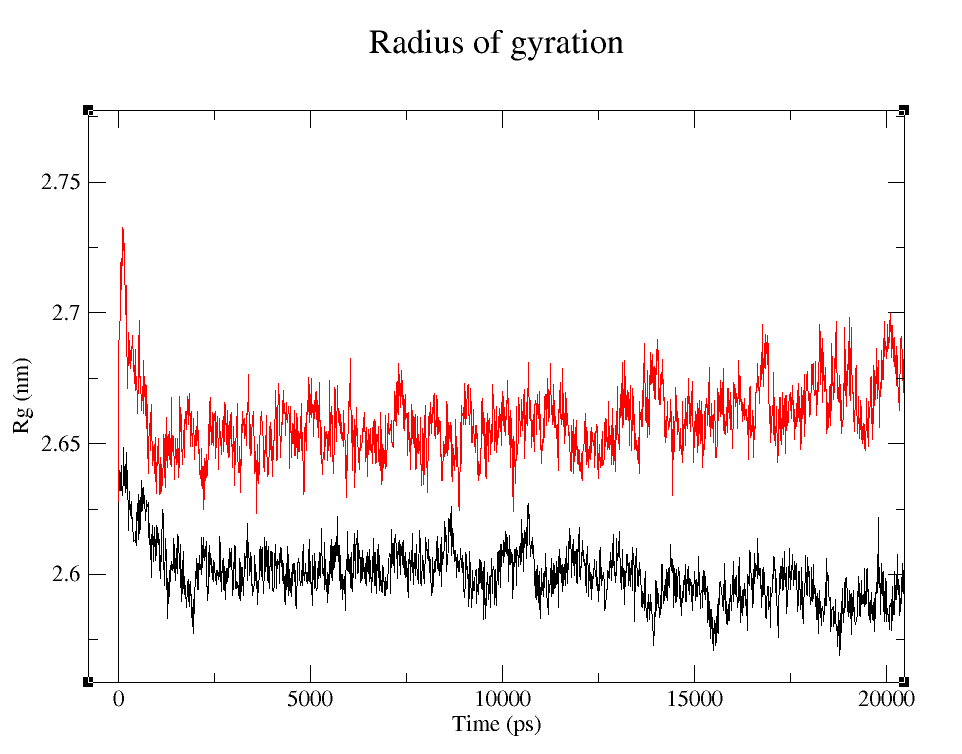
- 1) Comparing Radius of Gyration (Rg) of the WT and modified proteins through ~90 ns simulations (Figure 4).
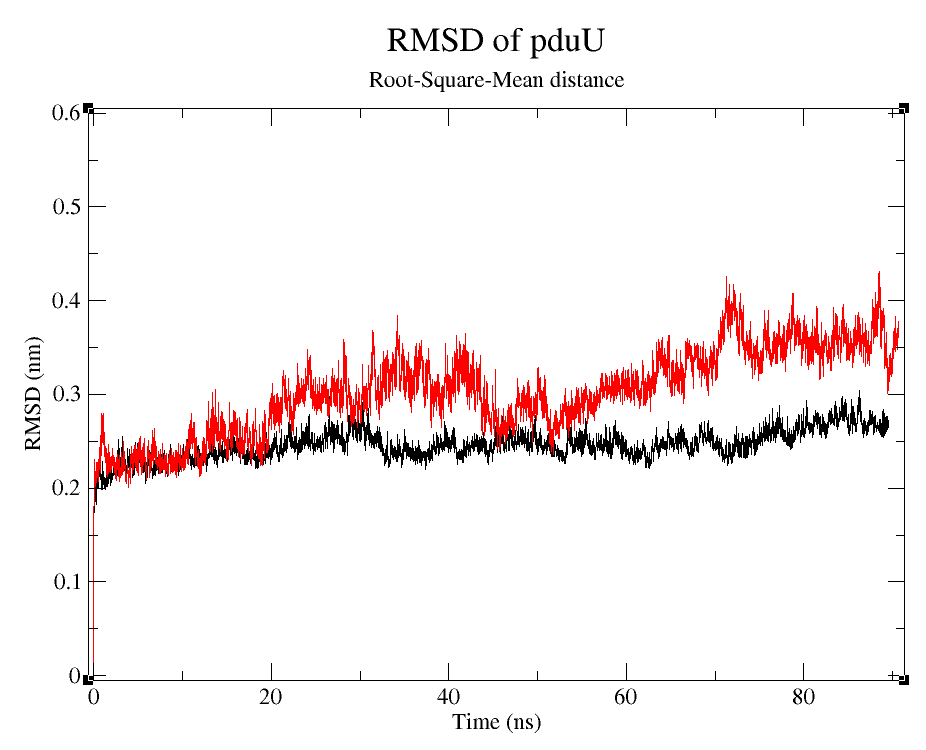
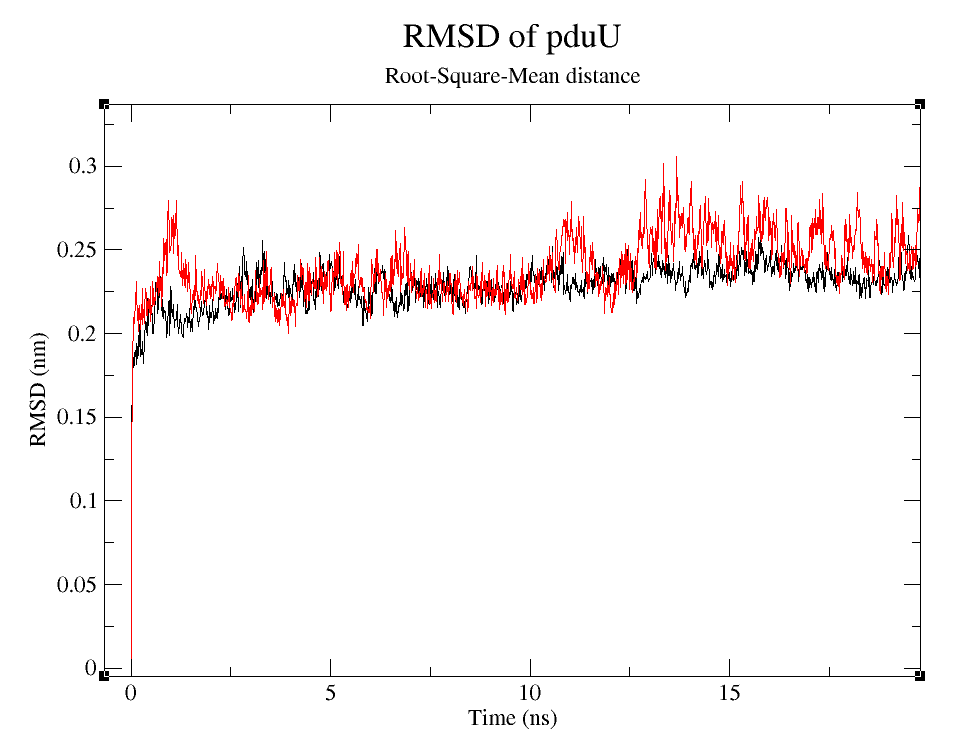
- 2) Comparing Root-Mean-Square deviation (RMSD) of the WT and modified proteins through ~90 ns simulations (Figure 6).
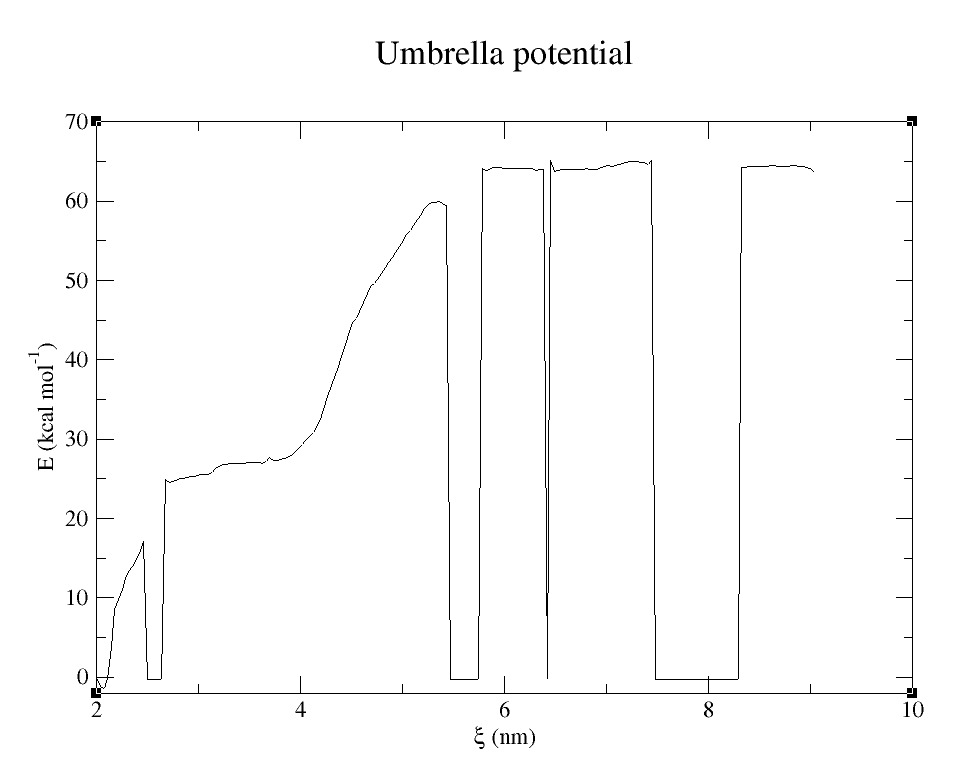
- 3) Comparing free energy of dimmer formation of both proteins using Umbrella Sampling (US) technique (2). See Figure 8.
Radius of Gyration
Radius of gyraton is a measure of how apart is the mass of an object distributed around it's center of mass.
It's easy to see that the average folding of both, wild type and modified proteins, is maintained throughout the whole simulation. The fact that the Rg is bigger for pduUmod means that the modification allowed the whole protein to increase in volume, i.e. it is a little less tightly folded than pduUWT, but since the value reached is maintained throughout the whole simulation, we can infer that the modification did not affect the stability enough to make it unfold.
Root-Squared-Mean distance
RMSD is often used to prove stability of preteins numerically (Kumar 'et. al.' 2014).
We can observe that, despite the fact that pduUmod RMSD is a little bigger than pduUWT RMSD, both are still pretty stable during the simulation. Usually, those changes in RMSD wou
Free Energy: dimmer formation
Umbrella Sampling technique can be used to study and analyze the free energy of formation of proteins, ligand and protein complex, binding of two molecules, etc.
Because of lack of computational time at the Super Computing Center, we could not complete the Potential Mean Force for the pass of the molecule though the porous. Result might be shown in Jamboree presentation.
Permeability
We also needed to verify that the new porous allowed the substrate of the metabolic pathway to pass through the proteinic shell of the BMC. Which we also did using Umbrella Sampling technique.
Because of lack of computational time at the Super Computing Center, we could not complete the Potential Mean Force for the pass of the molecule though the porous. Result might be shown in Jamboree presentation.
References
¹ Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., Heer, F.T., de Beer, T.A.P., Rempfer, C., Bordoli, L., Lepore, R., Schwede, T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46(W1), W296-W303 (2018).
² Hub JS, Groot BLD, Spoel DVD. g_wham—A Free Weighted Histogram Analysis Implementation Including Robust Error and Autocorrelation Estimates. Journal of Chemical Theory and Computation. 2010;6(12):3713–20.
³ Jorda, J., Liu, Y., Bobik, T. A., & Yeates, T. O. (2015). Exploring bacterial organelle interactomes: a model of the protein-protein interaction network in the Pdu microcompartment. PLoS computational biology, 11(2), e1004067. doi:10.1371/journal.pcbi.1004067
⁴ Humphrey, W., Dalke, A. and Schulten, K., "VMD - Visual Molecular Dynamics", J. Molec. Graphics, 1996, vol. 14, pp. 33-38.
⁵ H. Bekker, H.J.C. Berendsen, E.J. Dijkstra, S. Achterop, R. van Drunen, D. van der Spoel, A. Sijbers, and H. Keegstra et al., “Gromacs: A parallel computer for molecular dynamics simulations”; pp. 252–256 in Physics computing 92. Edited by R.A. de Groot and J. Nadrchal. World Scientific, Singapore, 1993.
⁶ Kumar, C. V., Swetha, R. G., Anbarasu, A., & Ramaiah, S. (2014). Computational Analysis Reveals the Association of Threonine 118 Methionine Mutation in PMP22 Resulting in CMT-1A. Advances in bioinformatics, 2014, 502618. doi:10.1155/2014/502618