Contents
INTEGRATED BIOCONTAINMENT CONSTRUCTION
Introduction
Planned implementation of the project
We, as a team, believe that our project is a viable alternative to the use of petroleum-derived compounds, and we envision our idea being used in industries to re-purpose plant-derived waste into valuable chemical products. We believe that it is crucial to think ahead about the problems that the implementation of our project may face in the future. However, this raises the concern on how are we going to ensure that in the future, our organism remains contained within the industrial facilities where it is handled. Now-days, bioreactors are the most standardized way in which chemicals are produced with the use of industrial biotechnology, and their use under the right conditions allows to handle GMOs safely; therefore the bio-containment strategy focus on the implementation of the GMO in one of these devices. The most likely scenario in which our organism may be accidentally released is during the operation and maintenance of the bioreactor, in which small volumes of the media in which the bacteria are being grown may escape as an aerosol or drops; however the possibility of an accident where the reactor is broken and its contents are fully spilled can not be dismissed.
Our bio-containment proposal: A light dependent strain
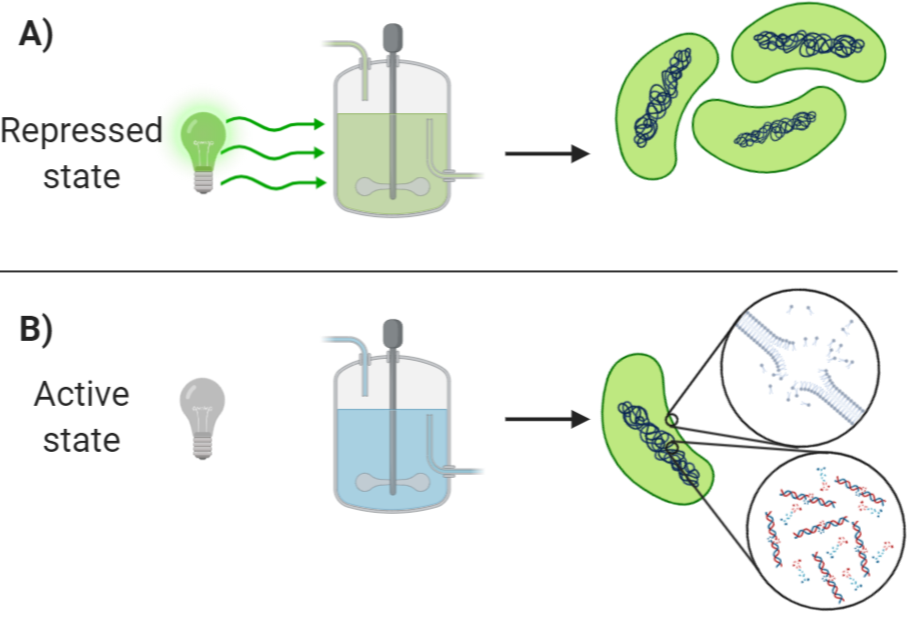
Nowadays, there are several ways to induce the death of bacteria that escape containment conditions; for example, there's auxotrophy where an organism can only live in the presence of a particular nutrient. For example, there are already commercial strains modified to be dependent of synthetic amino acids, or some others that are unable to synthesize vital components that have to be artificially added into the growth medium to keep them alive. However, this kind of biocontainment methods have their limitations, such as the costs of the substances that have to be added to keep the culture alive, or the fact that if there is an accidental spill or their container is broken. There's medium available in the floor. The bacteria can be kept alive for some time. In other words, there is not an "on and off switch" that can kill the bacteria, but there is more of a gradual decrease in the nutrient concentration instead of rapid death. Also, the use of these auxotrophic strains alone may not eliminate the DNA of the genetically modified bacteria, so there is a small chance of horizontal gene transfer to wild type of bacteria even after the original bacteria died. Our plan with this safety construction is to destroy the bacteria quickly, and ensuring that it does not survive outside containment and to destroy the DNA of the modified bacteria, to prevent gene segments that may be released into the environment.
To maximize the control of where can our organism can live, we propose the use of construction with the only purpose is to destroy the bacteria if it comes out of containment and by all possible means prevent its growth and proliferation of the organism outside containment. It was specially designed to avoid horizontal gene transfer and colony formation. This construction was assembled as an in-silico design only; since the main objective of the project was to process Vanillate into Muconic acid inside a bacterial microcompartment. However, we believe that this construction is feasible since the genes that compose it was tested in previous research projects.
The construction
The parts of the biocontainment circuit
| Name | Description | Type |
|---|---|---|
| <partinfo>K3317052</partinfo> | Serratia marcescens SM6 extracellular secretory protein (NucE) | Coding |
| <partinfo>K3317053</partinfo> | Putative phage lysozyme (NucD) | Coding |
| <partinfo>K3317062</partinfo> | Updated version of the Heme oxygenase 1, first 15 codons optimized (ho1*) | Coding |
| <partinfo>K3317063</partinfo> | Phycocyanobilin: ferredoxin oxidoreductase, fully codon optimized (pcya**) | Coding |
| <partinfo>K3317064</partinfo> | Constitutive promoter (PrpsD) | Regulatory |
| <partinfo>K3317066</partinfo> | Sensor kinase CcaS, first 15 codons optimized (ccaS*) | Coding |
| <partinfo>K3317069</partinfo> | Miniaturized sensor kinase miniCcaS#3, first 15 codons optimized (ccaSm3*) | Coding |
| <partinfo>K3317055</partinfo> | RBS designed by the safety team specifically for NucE overexpression to citotoxic levels in E.coli K12 | RBS |
| <partinfo>K3317056</partinfo> | RBS designed by the safety team specifically for NucD overexpression to citotoxic levels in E.coli K12 | RBS |
| <partinfo>K3317057</partinfo> | RBS designed by the safety team specifically for NucA overexpression to citotoxic levels in E.coli K12 | RBS |
| <partinfo>K3317058</partinfo> | RBS designed by the safety team specifically for φX174E overexpression to citotoxic levels in E.coli K12 | RBS |
| <partinfo>K3317068</partinfo> | Synthetic RBS in pMF35 | RBS |
| <partinfo>K3317072</partinfo> | Slightly modified version of the MF001 Synthetic RBS. | RBS |
How does it work?
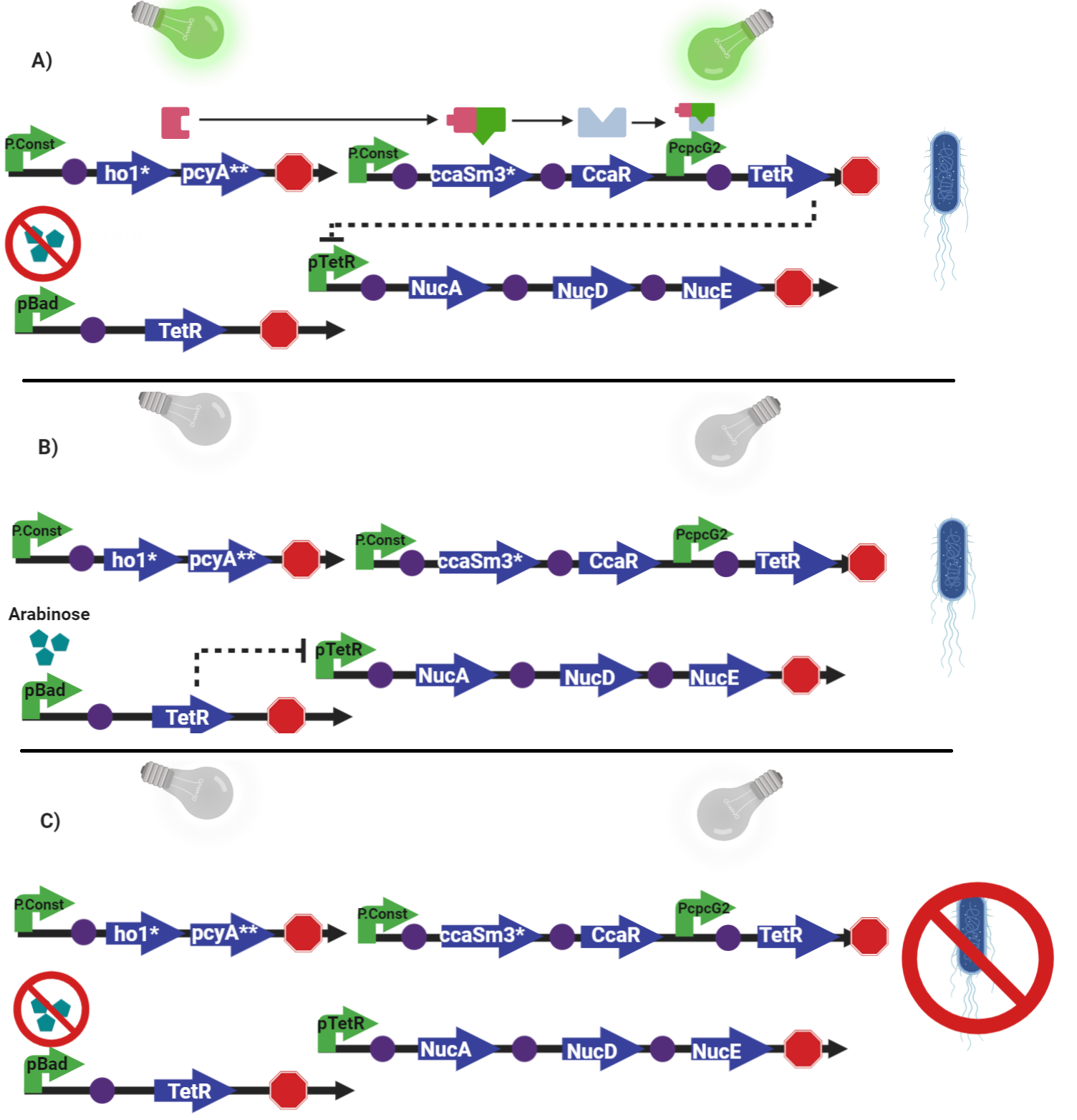
The backdoor circuit
To be able to keep the organism alive without the green light; for example during transport, quality management or inoculation of other growing media, a back-up system had to be added. This is a straightforward system activated by arabinose with the pBad promoter that expresses TetR whenever arabinose is present. In the case of our bacteria, to maintain the organism alive, arabinose should be artificially added into the medium when the green light is not present; however this backdoor circuit can be modified with other possitively regulated promoter other than pBad depending on the future applications of this part.
References
- Berkmen, M., Benedik, M., & Bläsi, U. (1997). The Serratia marcescens NucE protein functions as a holin in Escherichia coli. Journal Of Bacteriology, 179(20), 6522-6524. doi: 10.1128/jb.179.20.6522-6524.1997
- Castillo-Hair, S., Baerman, E., Fujita, M., Igoshin, O., & Tabor, J. (2019). Optogenetic control of Bacillus subtilis gene expression. Nature Communications, 10(1). doi: 10.1038/s41467-019-10906-6
- Chapter 14 Bioreactors. (2019). Retrieved 21 October 2019, from https://frtr.gov/matrix2/health_safety/chapter_14.html
- Jin, S., Chen, Y., Christie, G., & Benedik, M. (1996). Regulation of theSerratia marcescensExtracellular Nuclease: Positive Control by a Homolog of P2 Ogr Encoded by a Cryptic Prophage. Journal Of Molecular Biology, 256(2), 264-278. doi: 10.1006/jmbi.1996.0084
- Kill switches for engineered microbes gone rogue. (2019). Retrieved 21 October 2019, from https://wyss.harvard.edu/news/kill-switches-for-engineered-microbes-gone-rogue
- Merseburger, T., Pahl, I., Müller, D., & Tanner, M. (2013). A Risk Analysis for Production Processes with Disposable Bioreactors. Disposable Bioreactors II, 273-288. doi: 10.1007/10_2013_244