Contents
BIOSAFETY CONSIDERATIONS TO MINIMIZE THE RISKS INVOLVED IN THE PROJECT
Introduction
Legal Perspective
Mexico is a part of two major international agreements: one of them is the Cartagena protocol, while the other is the “Nagoya-Lumpur protocol on liability and compensation”. The Cartagena Protocol is an international agreement that deals with biosafety issues; this concluded within the framework of the Convention on Biological Diversity (CBD). The CBD has many more general purposes for the conservation and sustainable use of biodiversity and the responsible use of genetic resources. The function of the protocol is to guarantee an adequate level of protection concerning the transfer, management and use of living modified organisms of the current biotechnology application which may affect the responsible use of biological diversity and its conservation.
This alliance promotes different procedures to carry out safe handling, transfer and use of LMOs. This protocol proposes to analyze different previous criteria depending on the situation. These are based on the analysis of possible future effects of LMOs and evaluation of the extent of the damage if they are released before transfer. The Nagoya-Kuala Lumpur protocol is an international protocol which has the function of complementing the Cartagena protocol contributing to the conservation and sustainable use of biodiversity.
The evaluation of other factors, such as risk to human health, based on responsible international standards and procedures, is critical.
The objectives of this protocol are to seek to prevent, reduce, contain and avoid the possible damage caused by living modified organisms (LMOs).
Protection Objectives
- Human:
- Protecting the team’s safety in the work environment with an influential culture of prevention.
- Ensuring that every member of the team involved with laboratory work is qualified to handle the material safely and are also prepared to handle possible risk scenarios related to the development of the project.
- Protecting the community’s well-being by ensuring the isolation of the GMO and its genetic material, and also by avoiding the use of any pathogenic genes or species that may be harmful to humans, animals or plants.
- Environment:
- Ensuring that the GMO and its genetic material remains contained within the limits of the laboratory setting.
- Preventing the gene-leaking caused by horizontal gene transfer.
- Avoiding the use of any pathogenic genes or the handling of species that may be harmful to humans, animals or plants.
- Laboratory:
- Ensuring the appropriate use of the laboratory equipment by following the standardized protocols
- When possible, avoiding the use of harmful chemicals such as Ethidium bromide with safer alternatives such as GelRed; and when the use of a safer alternative is unfeasible, making sure that all of the preventive measures are being taken to prevent any harmful effects caused by the material
Assessment of the Possible Risks Associated with the project
Brief description of the project:
This project is based on the feasibility of using Bacterial Micro Compartments (BMC) as microreactors for industrial purposes to obtain phenolic compounds with the conversion of black liquor (BL) from the lignocellulosic waste of paper industry. This was achieved through synthetic biology by isolating a triple enzyme pathway (Protocatechuate decarboxylase, 3-O-methylgallate-O-demethylase and Catechol 1,2-dioxygenase) that converts Vanillate, a compound present in BL, into cis-Muconate, a chemical used in nylon production, inside of a modified BMC. By isolating the process through encapsulation, the enzymes within the BMC increases their efficiency by reducing the distance between them, while also decreasing the cytotoxicity of the compounds involved. Protein engineering was used to modify the structure of the wild type BMC, and molecular dynamics predicted their interactions as well as modelling the metabolic system. With this approach, our future goal is using modified BMCs as a device to optimize the biosynthesis of other compounds.
Description of the proposed organism
The present GMO consists of the insertion and cloning of construction, which contains both structural and a synthetic metabolic route. The metabolic component integrates a set of non-pathogenic gene sequences coding for enzymes originating from Pseudomonas putida mt-2, Sphingomonas paucimobilis SYK-6 and Klebsiella pneumonia. The structural component of the construction consists of and a modified structural gene sequence from Salmonella enterica LT-2. The construction was inserted into Escherichia coli K12 DH5alpha/Top10/BL21 using standard molecular biology techniques.
The parts used to construct the metabolic pathway to process the Vanillate into Muconic acid were chosen because they were used in a laboratory without any significant safety concerns associated with them. The parts used do not contain sequences that increase the virulence or any other risk factors in the recipient organism.
Purpose of the GMO
- General objective of the project:
- Designing an expression cassette that converts Vanillate into Muconic acid works as an isolated microreactor inside of an E.coli chassis.
- Short term objectives:
- Using synthetic biology techniques to create an enzymatic route that converts Vanillate into Muconic acid.
- Using protein engineering to modify the natural version of the structure of the PduU structural protein
- Expressing the modified version of PduU protein in E. coli
- Comparing the capacity of the modified PduU to improve the efficiency of the enzymatic route with the unmodified version.
- Expressing the enzymatic route inside of the microcompartment, and comparing its efficiency to
- Long term objectives:
- To develop a viable biotechnological way to convert lignocellulosic waste products into valuable aromatic compounds.
Identification of potential hazards of the GMO
The recipient organism:
The recipient organism is Escherichia coli K12 DH5α/Top10/BL21, which are commercially available. The strains derived from E.coli K12 are some of the most studied organisms in molecular biology, and they have been completely sequenced. Nowadays, because of its predictability and history of safe use, E.coli K12 variants are routinely used for studies in molecular biology as a model organism; therefore, it can be considered safe to work. Derivatives of K12 are considered a-virulent (Hacker and Ott, 1992) and may be classified in the CDC biological agents hazard group 1 because these strains have no adverse effects on human, animal or plant health or the environment, and there is an established record of safety in the laboratory with (Tian and Tao, 2014). Furthermore, the threeE.colistrains that were used (DH5α/Top10/BL21) are incapable of surviving outside the laboratory environment.
The donor organisms:
Note that NONE of the donor organisms was directly handled in the lab; only their sequence was used. The sequences of interest were sent to be synthetized by IDT
Pseudomonas putida mt-2:
- One of the donor organisms is P. putida is a highly characterized aerobic saprotrophic bacteria that inhabits most nutrient-rich water and soil. It is a rod-shaped, flagellated, gram-negative bacterium that grows optimally at 25-30 C (Anzai et al., 2000). According to the U.S. Public Health Service Guidelines it is t is classified as a level 1 safety hazard because it does not represent a significant risk towards the environment and the people exposed to it since it is not considered a primary pathogen, even though, there have been reports of isolated incidents of it affecting a human hosts (Yang et al., 1996). It was the first Gram-negative soil bacterium to be certified by the Recombinant DNA Advisory Committee of the United States National Institutes of Health as the host strain of host-vector biosafety (HV1) system for gene cloning in Gram-negative soil bacteria (Stjepandic, et al., 2002).
Sphingomonas paucimobilis SYK-6:
- Another donor organism is Sphingomonas paucimobilis, which is naturally present in soil, water and plants. S. paucimobilis are gram-negative bacilli non-fermenting, strictly aerobic, oxidase and catalase-positive which grow at room temperature. S. paucimobilis is the only species described in human infections from the family Sphingomonadaceae. Based on the U.S. Public Health Service Guidelines, it is categorized as a risk group 2 organism. It is an aerobic Gram-negative bacillus that, although rare in humans, it has been reported as an opportunistic pathogen which can infect immunocompromised and hospitalized patients. It has a low virulence and a very low mortality rate (Glaeser and Kämpfer, 2014).
Klebsiella pneumonia:
- Klebsiella pneumonia is Gram-negative, non-motile, usually encapsulated rod-shaped bacteria, belonging to the family Enterobacteriaceae. These bacteria produce lysine-decarboxylase but not ornithine-decarboxylase and are generally positive in the Voges-Proskauer test. They are generally facultatively anaerobic, produce mucoid colonies and range from 0.3 to 1.0 mm in width and 0.6 to 6.0 mm in length (Podschun and Ullmann, 1998). Based on the U.S. Public Health Service Guidelines, it is classified as a risk group 2 organism. K. pneumoniae is the most pathogenic strain to humans among all Klebsiella spp. It is a significant cause of primary liver abscess and microbial fascial space infections among diabetic patients in Asia, predominantly in Taiwan, it is commonly isolated from infections affecting burns and human bites. It recently has become an increasing cause of chronic diarrhoea in HIV infected adults in Africa.
Salmonella enterica LT-2:
- Salmonella enterica subspecies enterica has 2610 different serotypes, the most well known being serotypes Typhi, Paratyphi, Enteriditis, Typhimurium and Choleraesuis. Salmonella enterica is a facultative anaerobe and is a gram-negative, motile and non-sporing rod that is 0.7-1.5 by 2.0-5.0 µm in size (Public Health Agency of Canada, 2010). All Salmonella enterica subspecies (except for serotype Typhi) are found in blood, urine, faeces, food and feed and environmental materials (Gianella, 1996). Salmonella enterica is classified as a risk 2 microorganism according to the U.S. Public Health Service Guidelines. Salmonella enterica can cause four different clinical manifestations: gastroenteritis, bacteremia, enteric fever, and an asymptomatic carrier state. It is most common in children under the age of 5, adults 20-30-year-olds, and patients 70 years or older.
The insert:
- Inserts constitute structural gene sequences encoding modified bacterial micro-compartmentalization genes, and non-structural genes encode enzymes and signal peptides from different organisms, as a part of a novel integrated metabolic pathway. These enzymes were expressed at high levels in E.coli to obtain enough product for functional analyses.
- The insert also includes a modified version of the PduU structural protein to form the internal micro-compartments.
Toxicity of recombinant proteins
- None of the enzymes (3-O-methyl gallate-O-demethylase, Protocatechuate decarboxylase, and Catechol 1,2-dioxygenase) and and structural proteins (Pdu) that were used for this project are known to be involved in pathogenicity of their hosts, however they do play physiological roles in their native hosts.
Vectors used:
- The vectors used for this project are plasmids supplied by the iGEM organization. Both are pUC derived, and they do not possess severe pathogenic traits. However, they do present antibiotic resistances, pSB2K3 to Kanamycin and pS1C3 to Chloramphenicol, these resistances are for selection purposes and do not represent a risk.
The resulting GMO:
- The final product consists in a non-pathogenic E. coli chassis with an integrated system that functions as a micro-reactor composed of a network of three enzymes encapsulated inside a proteinaceous micro-compartment. This construction allows the bacteria to process Vanillic acid into Muconic acid.
Concentration and scale of the project:
- For the experimental phase of the project, the GMO was cultured on LB solid media (20 Petri dishes at a time). The bacterial stock was stored and handled in 50 µl tubes.
Culture conditions of the organism:
- In the experimental phase, the GMO was incubated at 37°C for 24 hours in LB broth with the addition of vanillic acid. These activities took place in a certified Level 1 BSL laboratory located in the facilities of the UANL. No wind currents within the lab that may cause aerosols to spread in the room and all of the equipment has its designated area. The techniques that involved the handling of the organisms, like inoculation in medium and agar plates were in close proximity to a Bunsen burner. All of the disposable used tools and plates were sterilized before appropriate disposal.
Initial classification of the GMO:
Escherichia coli K12 DH5α /Top10/BL21 has a history of being a safe organism in the laboratory and therefore is unlikely to cause illness to humans, animals or plants. Also, genetic modifications are not expected to alter this characteristic. The pUC derived vectors also have a history of safe use and were provided by the iGEM Organization. Also, the inserts did not give the recipient organism characteristics that are likely to cause disease or to have ill effects on the environment. Therefore the GMO can be classified as a Risk Level 1 organism, which means that the risk of the occurrence of harmful effects is very low.
Formulation of the Problem and Hazards List
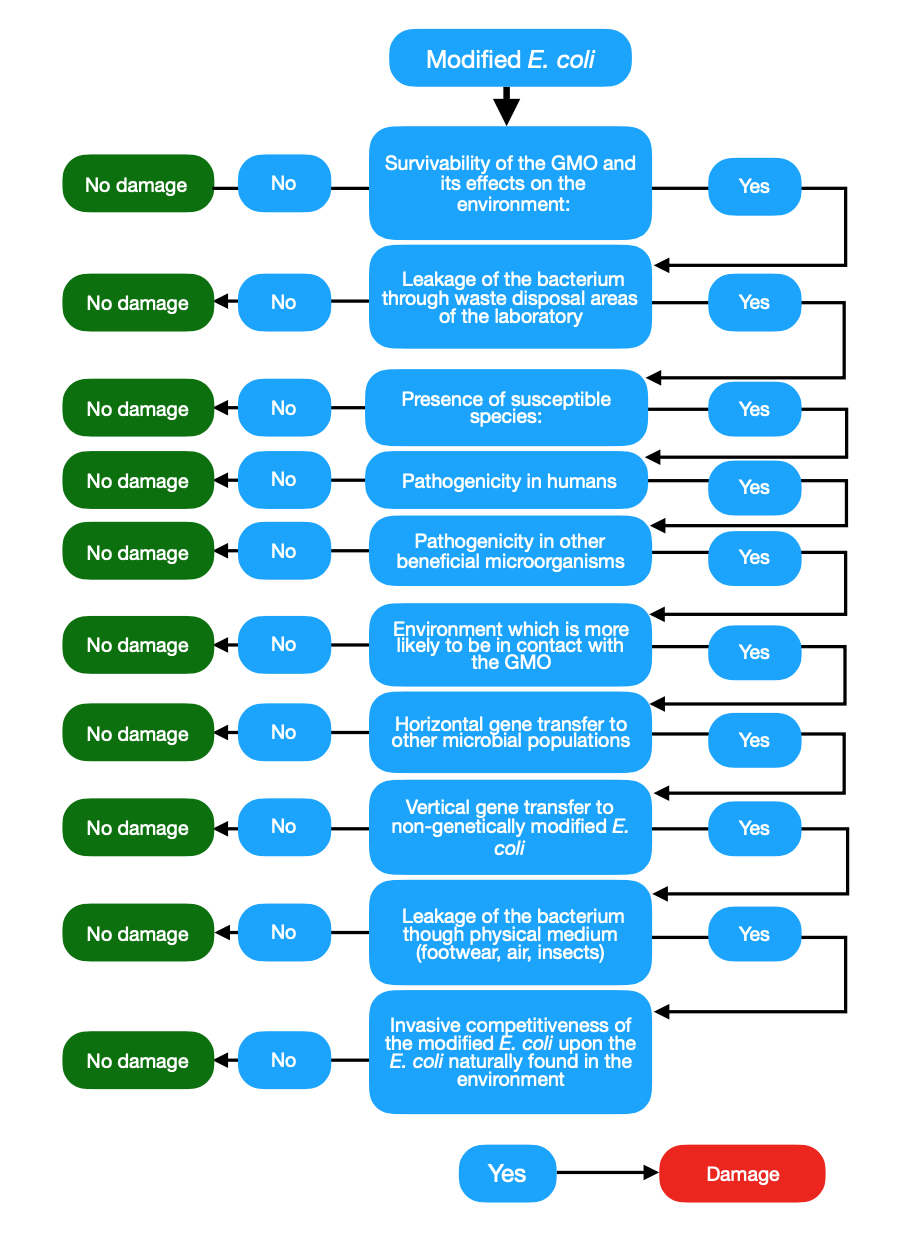
The possible hazards associated with the protection objectives of the modified E. coli bacteria under the environmental parameters could be:
1. Survivability of the GMO and its effects on the environment:
- a Leakage of the bacterium through waste disposal areas of the laboratory
2. Presence of susceptible species:
- a Pathogenicity in humans
- b Pathogenicity in other beneficial microorganisms
3.Environment which is more likely to be in contact with the GMO:
- a Horizontal gene transfer to other microbial populations
- b Vertical gene transfer to non-genetically modified bacteria
- c Leakage of the bacterium though physical medium (footwear, air, insects)
- d Invasive competitiveness of the modified E. coli upon the E. coli naturally found in the environment
Risk characterization
Risk of the activities that were involved in this project:
The only living microorganisms that were handled by the team are non-pathogenic strains of E. coli K-12 with low survivability outside laboratory conditions; therefore, the biological risk is very low. The experiments are standard molecular biology techniques, and the team is in compliance with iGEM’s safety and security rules and policies and is supervised at all times by at least one of the three instructors of the team. There is a subdivision within the team explicitly dedicated to monitor and to assess any risks involved in the project. The most important risk that was considered during the development of this project was the handling of pyrocatechol, which is a reactant with acute toxicity if in contact with skin, swallowed or inhaled. In order to mitigate the risks related to the handling of this substance,
Possible risks related to the chemicals associated with the GMO:
The two main chemical components that are going be involved are the Vanillic acid, as the input to the system and the Muconic Acid as the final product.
- Vanillic acid is described by the manufacturer as a not hazardous substance or mixture. It is not classified as a carcinogen by the ACGIH, NTP and OSHA at high concentrations.
- Muconic acid at full concentration is classified as an eye, skin and respiratory tract irritant agent.
Other reactants that should be taken into consideration:
- Pyrocatechol: It is a category 3 chemical, which is toxic if swallowed, digested, inhaled, or if it comes in contact with the skin. It also causes skin irritation and possible allergic reactions, along with eye damage, and it is suspected of causing genetic damage.
Environment which is more likely to be in contact with the GMO: During the experimental phase, the only environment that is at risk of being exposed to the GMO is the laboratory; however, it is not expected for the organism to survive outside containment. (Miyanaga K, Unno H, Tanji Y. 2006)
Presence of susceptible species: Neither of the three strains of Escherichia coli that were used as recipient (K12 DH5 ∝ /Top10/BL21) represents a risk towards humans, animals or plants; and the expressed gene products of the inserts are not considered harmful or capable of altering the pathogenicity/survivability/fitness of the recipient organism.
Survivability of the GMO and its effects on the environment: The strains of E. coli K12 that were used in the experiments have an extremely low survival rate outside containment (Miyanaga K, Unno H, Tanji Y. 2006). They also lack pathogenicity, therefore lacking the ability to affect the immediate physical environment.
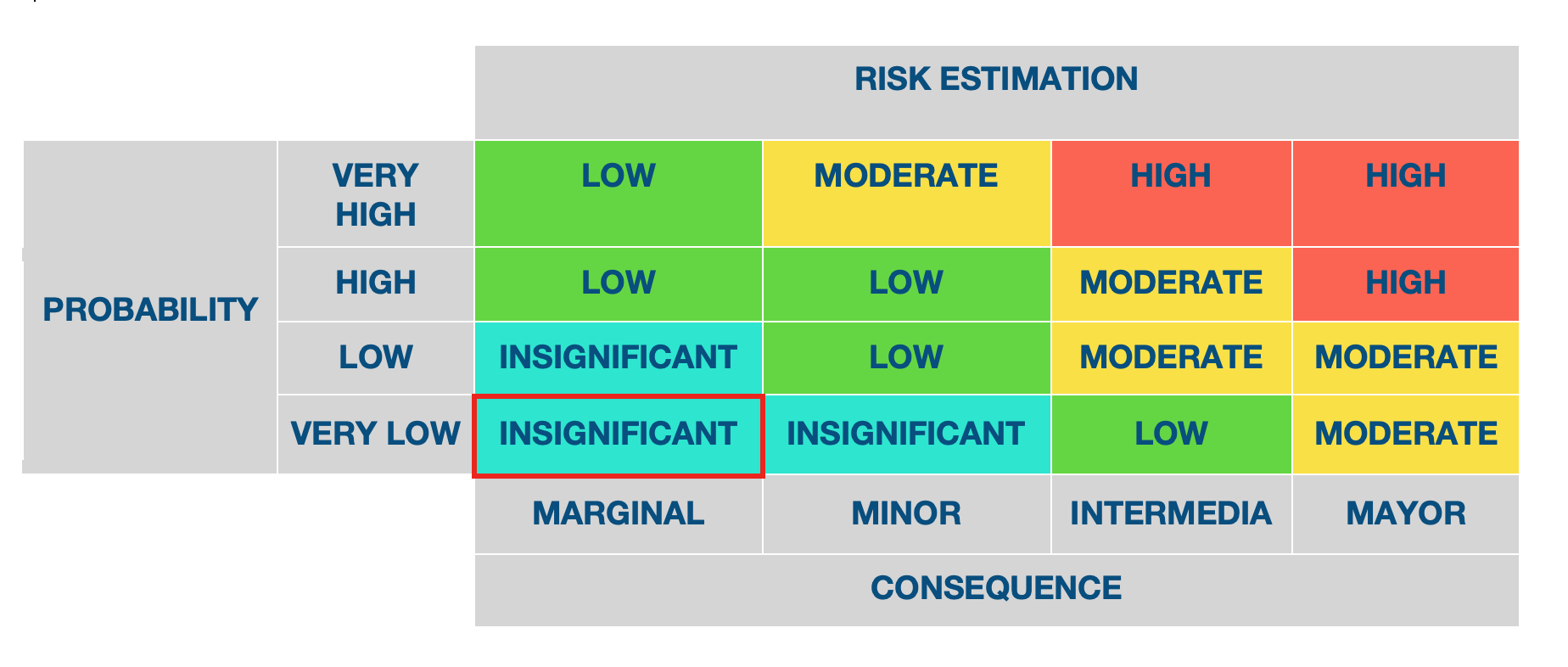
Risk Estimation
- Based on the list of hazards created in the last step, we can proceed to the study of binomial probability/effect of exposure for the estimation of risk in the previous context established (Figure 1 Path to damage). The following considerations are part of the Path to damage. Biologically, according to the history of safe use, the modified bacteria does not represent any hazard based on your safety record. Genetic transfer flow, pathogenicity and mutations, are therefore classified as very unlikely. With this, we can conclude that the estimated risk is negligible.
Risk Management
Safe lab practices
- Dra. Lydia Guadalupe Rivera Morales, is the executive secretary of the faculty's BioSafety committee, which is the organism that overlooks all of the research that takes place in this institution, managing biosafety-concerning issues at the institutional level; however, the team has taken its measures in a smaller scale by creating a team sub-division focused in the safety of our project.
- Everyone involved in any work in the laboratory must have gone through a training of the do's and don'ts in the lab before getting involved in the work
- At all times in the laboratory, the students involved in the team will be supervised by at least one of the instructors; and only the people who have had previous training in the use of a lab equipment and safety protocols will be allowed to perform experiments.
- Read our Laboratory rules
Waste treatment procedures
- Accidental spillages were dealt with disinfectant 10% Ethanol.
- Contaminated, solid waste such as plastic disposables, are bagged and autoclaved at 121°C for 20 mins, 1 bar pressure. The autoclave is located in the laboratory installations and is validated regularly. After being autoclaved, the waste is stored in a sealed in bins, with biohazard symbols displayed, until removal by a registered waste contractor (Nombre de la Empresa) for disposal.
- Sharp material was disposed of in cin-bins, which are removed by a registered waste contractor (Nombre de la Empresa) for disposal.
Final containment measures
- During the experimental phase of the project, the organism was contained inside a freezer of a level 1 biosecurity laboratory inside the UANL’s School of Biological Sciences facilities. When not in use, for storage purposes the culture was frozen and left at -80 Celsius.
- We designed a circuit dedicated only for Biocontainment purposes. For its construction we took into consideration the future applications of our project in the real life industrial scenario, and proposed a method to reduce the risk of the organism escaping with a light activated killswitch.
Bibliography:
- Anzai; Kim, H; Park, JY; Wakabayashi, H; Oyaizu, H; et al. (Jul 2000). "Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence". Int J Syst Evol Microbiol. 50 (4): 1563–89. doi:10.1099/00207713-50-4-1563.
- Hacker, J. and Ott, M. (1992) Pathogenicity testing. In: Safety in Industrial Microbiology and Biotechnology (Collins, C.H. and Beale, A.J., Eds.), pp. 75–92. Butterworth-Heinemann, Oxford.
- Glaeser S.P., Kämpfer P. (2014) The Family Sphingomonadaceae. In: Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F. (eds) The Prokaryotes. Springer, Berlin, Heidelberg
- Giannella RA (1996). Baron S; et al. (eds.). Salmonella. In:Baron's Medical Microbiology (4th ed.). Univ of Texas Medical Branch. ISBN 978-0-9631172-1-2.
- Peter Kuhnert, Patrick Boerlin, Joachim Frey, Target genes for virulence assessment of Escherichia coli isolates from water, food and the environment, FEMS Microbiology Reviews, Volume 24, Issue 1, January 2000, Pages 107–117, https://doi.org/10.1111/j.1574-6976.2000.tb00535.x
- R. Podschun, U. Ullmann (1998) Klebsiella spp. as Nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and Pathogenicity Factors Clinical Microbiology Reviews, 11 (4) 589-603; DOI: 10.1128/CMR.11.4.589
- Na SH, Miyanaga K, Unno H, Tanji Y. 2006. The survival response of Escherichia coli K12 in a natural environment. Appl Microbiol Biotechnol 72:386–392. doi:10.1007/s00253-005-0268-3.
- Salmonella enterica spp (formerly Salmonella choleraesuis). (2010). Retrieved from https://www.msdsonline.com/resources/sds-resources/free-safety-data-sheet-index/salmonella-enterica-spp/.
- Stjepandic, D., Weinel, C., Hilbert, H., Koo, H. L., Diehl, F., Nelson, K. E., … Hoheisel, J. D. (2002). The genome structure of Pseudomonas putida: high-resolution mapping and microarray analysis. Environmental Microbiology, 4(12), 819–823. doi: 10.1046/j.1462-2920.2002.00313.x
- Tian, Deqiao, and Tao Zheng. “Comparison and analysis of biological agent category lists based on biosafety and biodefense.” PloS one vol. 9,6 e101163. 30 Jun. 2014, doi:10.1371/journal.pone.0101163
- van Elsas, Jan Dirk et al. “Survival of Escherichia coli in the environment: fundamental and public health aspects.” The ISME journal vol. 5,2 (2011): 173-83. doi:10.1038/ismej.2010.80
- Wayne Parrott. María Mercedes Roca -- 1. ed. Guia para la evaluación de riesgo ambiental de organismos genéticamente modificados / [editores] Paulo Paes de Andrade, -- Sâo Paulo : Internacional Life Sciences Institute do Brasil, 2012.
- Yang C. H., Young T., Peng M. Y., Weng M. C. (1996). Clinical spectrum of Pseudomonas putidainfection. J. Formos. Med. Assoc. 95 754–761. [PubMed
