Model Analysis
Bifurcation analysis
Bifurcation analysis is a type of analysis which allows to understand how changes in values of one parameter affect the number of steady states of the system and their stability. Briefly, for each parameter value we can draw dx/dt over x graph and find the values x* at which the graph crosses the horizontal line (dx/dt=0), corresponding to steady states. After obtaining these x* values, we plot them in a graph where the horizontal axis represents a range of parameter values and the vertical axis represents the obtained steady state values. Systems having two steady states typically result in a bifurcation diagram with a distinct "saddle-node" shape.
P2 genetic switch
We ran the bifurcation analysis of the influence of one of the parameters, u, on the P2 genetic switch system and obtained the following diagram.

From the graph we can see that the selected parameter range does not yield a saddle-node bifurcation diagram, meaning that these parameter values do not give the system two steady states. It could be that other selected parameters (σ, γ, τ) also contribute to this result. This leads us to conclude that the P2 genetic switch system is highly sensitive to specific parameter values, which are fine-tuned in order to achieve the bistability, which is essential for the P2 bacteriophage to switch from lytic to lysogenic state and vice versa.
Sensitivity analysis
Sensitivity analysis allows to explore which parameters have the biggest influence on the system. We implemented sensitivity analysis on our models using SimBiology sensitivity calculation tool.
Model Plasmid
The figure below is the sensitivity analysis of C protein. As can be seen from the figure, C protein is sensitive to the amount of unbound DNA (D0 species), polymerase (P) and the amount of arabinose in the reaction. These species have the highest effect on the C protein. Parameters which also show to play a significant role in the behaviour of C protein dynamics are k0, which is the rate constant for C protein degradation, as well as k1 and k2, forward and reverse reaction rates for arabinose binding. It is no surprise that C protein degradation constant has the highest impact among all parameters involved as it is the only irreversible reaction that removes C protein from the system. The C protein behaviour also depends on kt1, C protein synthesis rate constant, as well as k4 and k5, C protein dimerization constants.

Here, we present the sensitivity analysis of the Cox protein in Model Plasmid. From the figure below, we conclude that similarly to C protein, Cox protein depends mostly on unbound DNA species, polymerase and arabinose concentration. Significant parameters include k1 and k2, forward and reverse reaction rate for arabinose binding, and k02, the degradation rate constant of Cox protein. kt2, reaction rate constant for Cox protein synthesis, also effects Cox behaviour.

These results give us insights into ways to better control the behavior of our systems. For example, the high influence of degradation rate constants on proteins informs us that the most effective way to manipulate protein dynamics could be by increasing or decreasing their stability. This could be achieved, for example, by changing sequences of the protein encoding genes on plasmids. In addition, we observed a strong influence that arabinose has on our system, in particular, its binding to AraC protein. The vector which we initially chose for our system does not encode for AraC protein. Based on these results, we made a suggestion to the wet lab team to change to a vector encoding for AraC protein. Having it on a plasmid would enable us to manipulate its sequence and hence, the arabinose binding constant.
Switch Plasmid
The sensitivity analysis for the Switch Plasmid is depicted in the three figures below. C protein is seen to be dependent on the constant for C protein degradation, k0, also somewhat on kt3, the reaction rate of slowed synthesis of C protein due to TetR binding, k22, reversible reaction rate for TetR binding at high concentration and k33, reversible reaction rate for arabinose binding to D0 (unbound DNA). Other influential species on C protein behaviour were D0 (unbound DNA), arabinose (A0), TetR (r) and TetR dimers (q).
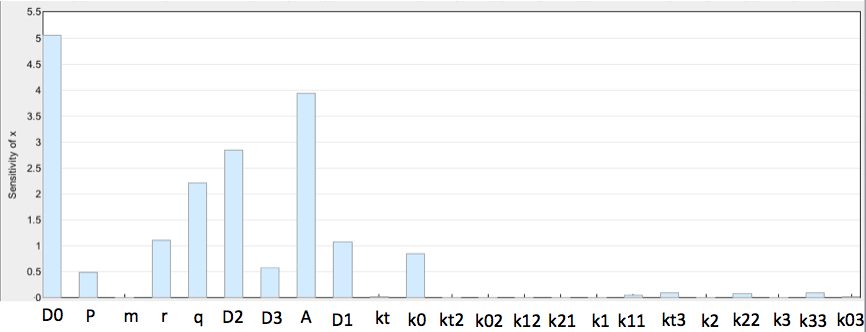
Cox is shown to depend primarily on D0, TetR, TetR dimers, D2 (DNA bound to TetR), D1 (DNA bound to arabinose) and arabinose concentrations. Out of all the parameters, it depends significantly on k02, degradation constant of Cox protein. k33, constant for arabinose biding on D0 species (unbound DNA), is also seen to have some effect on Cox protein.

TetR shows a similar trend to Cox: it depends mainly on D0, TetR dimers, D2, D1 and arabinose concentrations. However, we see an influence of TetR degradation constant (k03), arabinose binding constant (k33), constant for TetR binding in low concentration to DNA (k21), constant for TetR binding in high concentration to DNA (k22) and constant for TetR dimerisation reaction (k11) on its behaviour to a certain extent. Each of these have a variable effect on the behaviour of C protein within our system.

While we do not see the high sensitivity of this system to arabinose binding to various species as in the Model Plasmid, the degradation rates of the proteins still play an important role in this system's behaviour. Again, this can be controlled by regulating the stability of the proteins involved, in order to increase or decrease the rate at which they are degraded.

