Software
Software
Software
Motivation
Recent advancements in metagenomics suggest that current metagenomes hold the untapped potential to provide humanity with new drugs, antibiotics, industrial enzymes, and even more. However, designing functional metagenomic screens is a labor-intensive and time-consuming process, the essential part of which is knowing where to look. Extremophiles with enzymes that have biotechnological applications can be lurking anywhere from hydrothermal vents to permafrost or vulcanic regions. Thankfully metagenomic sequences from these and many more environments are being extracted at a faster pace and cheaper cost than ever before, therefore creating terabytes upon terabytes of information. Unfortunately, due to the complicated analysis pipeline and sophisticated software used for it, these data goldmines have been locked away from the public, and more importantly, researchers. Considering the complexity of this field and the amount of knowledge it requires, it would seem natural that these issues remained unchallenged, but solving them would mean unlocking these data mines and quite possibly marking a beginning of a new era in metagenomic research.
Vilnius-Lithuania 2019 team proposes a novel approach to the in-silico metagenomic research that could change the way we used to think about metagenomics. We managed to tackle the problem from its inner-heart and came up with a solution that is innovative and offers the approach to the variety of problems metagenomics holds. Our software provides an easy-to-use interface and a possibility to extract enormously rewarding results. It takes advantage of MG-RAST database API and allows users to search & download hundreds of thousands of metagenomic samples to our servers with just one click in their web-browser. Then it automatically standardizes and cleans the selected sequences & prepares them for further analysis. The user can choose from a variety of pre-made protein profiles obtained from the Pfam database; the primary tool used for similarity search is HMMER. Thus, the software allows anyone with only a basic understanding of IT to contribute to metagenomic research and advance science to the next level.
Tackling the problem
The primary objective of our IT team was to screen through metagenomic databases for light-inducible protein sequences for our wet lab team. For this task, we chose to use a popular tool for this job – Hidden Markov Models (HMMER) – a hmmer profile search against a sequence database. We chose to download our sequences from MG-RAST – currently the largest and professionally maintained metagenomic database. The first problem we encountered was gathering the sample sequences from the database - we discovered that accessibility, even for public data, is quite limiting. For example, we were unable to download the data in massive amounts. Therefore, we went to consult an experienced researcher in this field – Dr. Kazlauskas. We found that his team had the same issue with the same database – MG-RAST, and the bottom line was that they were unable to perform such downloads as well. During the discussion, we learned that even the downloading tool (which we built later on) would be a valuable asset to researchers in this field. We saw that the need for such software was imminent. This was the beginning of our journey towards Big-Seq software.
It didn't take long for us to discover that the data gathering problem was just the tip of the iceberg, and underneath lay much bigger problems. We discovered why performing such analysis was so hard, and the few scientific articles that mentioned MG-RAST didn't offer us any promising solutions as well. Specific needs for protein discovery analysis:
- Getting to know a selected metagenomic database takes time, especially if we intend to use its application programming interface (API). The majority of them do not offer convenient tools to download the data.
- Messy data – varying standards and dozens of formats brings confusion and makes it hard to adapt software settings to every kind of metagenomic data.
- Storage space – usually a personal computer does not have the extra few terabytes of space to store the downloaded data, and it is also a considerable problem for academic research since computers are usually not fit with this type of storage capabilities.
- Processing power – the processing requirements for tools like our sequence extractor and indexer are usually found in server-grade machines specifically built for these kinds of tasks.
- Knowledge of particular tools (HMMER, BLAST, mmseqs.) - it takes weeks of testing, learning, and configuring the software to work just the way you intend it to work, and for someone entirely new to this, it may seem repulsive and time-consuming.
- Programming knowledge & experience – since the data is varying from one database to another, the researcher will almost inevitably encounter a problem, like conversion or taking out an unwanted character. These tasks require some decent scripting knowledge - not a thing one can learn on-the-go while also being an absolute must in this field.
- Bioinformatics knowledge (profile building, file readings, formats) - a general understanding of what is a fasta format, why would you need to press a profile, error handling, and many more topics researchers would encounter throughout such and similar analysis.
- Linux operating system – since our primary analysis tool - HMMER may have a version for the Windows operating system, the complimenting applications usually don't. In most cases, they are built for Linux operating systems, so it is a consensus to work in a Linux environment for this type of analysis.
The solution
Our state of the art software features a user-friendly interface, self-explanatory protein discovery pipeline, while at the same time providing a Pfam's protein profiles for users to choose from.
The whole protein discovery pipeline involves several steps. First, gathering the metagenomic data to search proteins in. We took advantage of the MG-RAST database API and built a more convenient search engine & data extraction tool that can download huge volumes of data with little to no effort. This approach meets the needs of every student, scientist, or enthusiast who wants to analyze the metagenomic data but is unable to due to complex reachability.
After the data is downloaded, the next step consists of cleaning and standardizing the data. We came up with a solution to a well-known "duplicates and residues" problem, which could also solve similar problems in the big-data science field. Our solution enables users to prepare their data for any tool and analysis by cleaning and standardizing it. The output data can be used for further analysis with other popular tools.
The last step involves a protein profile scan against a sequence database. Sophisticated yet effective, open-source tool HMMER and its minapps are now made to work in the background without the users' need to set the complicated options in the terminal for each task - this eliminates the need to spend countless hours of reading the manual. The optimal computational power is also provided within our servers, enabling users to perform analysis on any machine that has access to the internet.
How it works
Type any desired keyword of choice, for example, a biome name – water in the search bar and allow the engine to display the search results.

At this point, our server makes a request to the MG-RAST API with users keyword and searches in their database - if the keyword finds the associated data, it downloads automatically to our server. Files should display in the results section on our website.

After the data downloads, you are asked to select the protein profile from the given drop-down menu. The menu stores the majority of popular protein profiles from Pfam.

The last step is to analyze the data – at this point, everything happens "behind the curtains." The downloaded data is scanned for duplicate sequences, and if any was found, they get removed from the original file. Also, the sequence file goes through "standardization," which removes the illegal characters and sequences that were too short. After that, the files are ready for HMMER analysis, which searches for similar proteins (according to selected profile) against your downloaded sequence database.
Output sequences show up at the bottom section of the website – these sequences are the ones that HMMER determined as similar to the profile of your choice.

Software integration
The fasta file format used in our software is a standard in bioinformatics and is recognized by popular tools, such a BLAST, HMMER, mmseqs, and many more. Output data can be used for further analysis with similar tools. Not only the input and output data is a well-known standard, but the HMMER software that runs in the background is one of the most popular tools for this kind of task.
Pfam
Pfam is a protein family database, which conveniently stores hmmer generated profiles. HMM, profiles are used as the description of the selected protein family to search against the sequence database. They are provided in our applications drop-down menu for users to choose from. They are essential for protein discovery analysis as they are the main focal point for HMMER software to compare sequences to.
Hidden Markov Models
Hidden Markov Models (HMMER) is a sequence analysis tool written by Sean Eddy. It is widely used in genomics for sequence search and perfectly serves our purpose while being more precise than other popular tools, such as BLAST. HMMER is a primary analysis software used in our pipeline as it scans through a sequence database and compares the sequences according to a selected profile.
Optimization tools
One of our great concerns was to find a way to remove duplicated sequences in large files from the initial data. Duplicates in the analyzed data can cause unexpected results, and most tools are not allowing such inconsistencies. So the removal of such duplicates is a must before we can continue with our analysis. One of the options we tried was to use a popular clustering tool – MMSeqs2 – a suggestion from our leading bioinformatics expect Dr. Justas Dapkunas. While being a prominent tool, it would still leave the duplicates in the output data. We discovered that it was a standing issue, so we decided to write our own algorithm, which would not only remove the duplicates but would clean the file from other residues. Our algorithm would allow us to standardize the file for specific tool requirements, in our case – HMMER. It is a long-needed tool for the scientific community, and it helps to advance metagenomics even further.
HMMER's requirements for specific tasks allowed us to mark the bounds of standards to our data. For example, the - hmmpress function required for profile database scanning has to be "pressed" before any further analysis can be performed. In this case, the protein profile has to have the right extension and prepared with the same version of HMMER software.
Code
The code is written in PHP, JavaScript, Bash, and Bootstrap framework. Everything is set o a Linux Ubuntu 18.04 LTS server.
Future improvements
Considering the fascinating applications of this software, we are currently planning future improvements. They are as follows:
- Clustering the most used data and maintaining it on our server for even faster analysis.
- The building of a passive protein-miner bot which would analyze the database in a long-time period and store the findings in the database as a mapping file, so that eventually we could have metagenomic protein indexing engine.
- For code improvements, we are planning to rebuild our application on more advanced languages like Go, React, Node, Python.
- Hardware improvements would include more storage, more memory, and higher processing power.
A potential
By providing users with our software, we give them the ability to find proteins no one has ever seen before, meaning there could be numerous drugs, antibiotics, enzymes, and much more protein compounds the world has never seen before, thus providing advancements in medicine, biology, and chemistry. With future improvements, it might become a cornerstone technology used in everyday biotechnology to look for novel parts in metagenomic samples.
Software Application
Metagenome-derived BLUF-cI: two domains for the precise modulation at the transcriptional level
Metagenomic DNA sequences open a unique opportunity to explore limitless environments harboring massive amounts of unknown and mostly unculturable organisms, including microbes. The breakout of high-throughput DNA sequencing and evolved molecular techniques made a significant impact on the bioactive molecule identification. However, determining the function of sequenced but uncharacterized genes remains a severe issue.
As a part of our project, we have created a software, allowing to mine for light-activated and DNA-binding domains containing proteins in unexplored metagenomic datasets. The gene coding for blue light-activated BLUF and DNA-binding cI repressor-like domains was encountered. By applying bioinformatic analysis on the metagenome-derived sequence, we were able to identified domains responsible for its characterization as light-activated DNA-binding protein.
Transcription is an essential step in gene expression, and its understanding has been one of the significant interests in molecular and cellular biology. By precisely tuning gene expression, transcriptional regulation determines the molecular machinery for developmental plasticity, homeostasis, and adaptation. The helix-turn-helix is a short motif found in a variety of transcription regulators 1. The domain is located in the N-terminal part of these transcriptional regulators. Its structure consists of about 50-60 residues, made up of two alpha-helices, connected by a turn. The second helix, known as the recognition helix, specifically interacts with the DNA. The two alpha-helices extend from the domain surface and constitute a convex unit able to fit into the major groove of DNA, where recognition helix makes most contacts with DNA 2.
Light is an essential factor that modulates the physiology of microorganisms. Three flavin-containing photoreceptor families are known to respond to blue light, and one of them is BLUF 3. BLUF domains are mostly found in prokaryotes, where they are found to be associated with a wide range of effector domains. They function as a light-activated switch, which tends to control gene expression or enzymatic activity, and this way modulates various physiological responses: phototaxis, nucleotide metabolism, biofilm formation, and others. BLUF domain contains ~100 amino acids. Based on structural studies of light-sensitive domain architecture, it suggests that it has ferredoxin-like fold with the FAD cofactor bound noncovalently between two parallel alpha-helical chains and surrounded by four essential amino acid residues 4. Blue light causes a rearrangement within the hydrogen bond network between the active site FAD and nearby conserved residues. Upon returning to darkness, the signaling state decays back to the ground state with kinetics that varies from seconds to minutes 5,6.
Design and experimental procedure
The idea behind BLUF-cI system was to use it as a transcription repressor. Since this metagenome-derived protein was not annotated, the first step of the experimental procedure was to identify its binding sequence. This was meant to be done by performing a few rounds of the systematic evolution of ligands by exponential enrichment (SELEX) method, followed by electrophoretic mobility shift assay (EMSA). After these experiments, BLUF-cI binding site would be identified by sequencing and used for BLUF-cI responsive promoter design (Figure 1).

Figure 1. Schematic representation of BLUF-cI responsive promoter design via experimental procedures.
Results
Team note: Working with novel, unannotated, metagenome-derived proteins might be challenging at times. This is what our team experienced while trying to achieve our goal – creating a BLUF-cI repressor based system for light-tunable transcription regulation.
The gene, coding for metagenome-derived BLUF-cI repressor protein, was chemically synthesized. For fast and efficient cloning procedure, the gene was inserted into pLATE expression vector and used for bacterial transformation.
Escherichia coli BL21 (DE3) cells, harboring the protein of interest, were induced with IPTG for BLUF-cI overexpression. SDS-PAGE analysis revealed the high yield of ~30 kDa protein in an insoluble form. To solve this problem, we thought of fusing our protein of interest to maltose-binding protein (MBP). However, the idea was declined since both protein termini have functional domains. Knowing this, we searched for methods, increasing the solubility of target protein, but not interfering with its functions. Thus we decided to use E. coli Arctic Express (DE3) strain, engineered to address the common bacterial gene expression hurdle of protein insolubility (Figure 3).
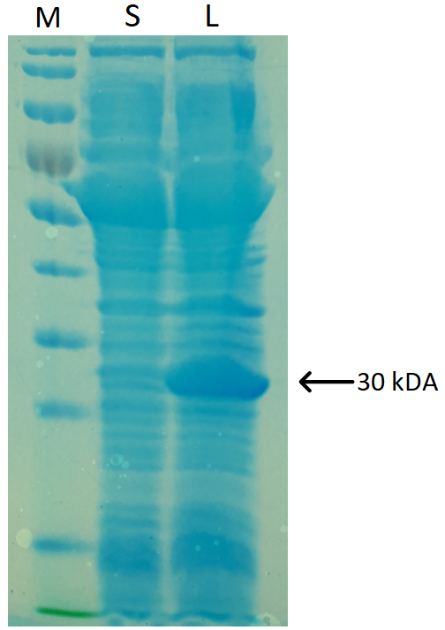
Figure 2. BLUF-cI SDS-PAGE analysis results. S – soluble protein fraction, L – cell lysate, M – PageRuler Prestained Protein Ladder. The arrow indicates the protein of interest.
To purify our protein for further DNA-binding analysis, various protein purification variants were tested. We started by performing a traditional immobilized metal ion chromatography (IMAC) technique, using Ni2+ or Co2+ ions, but because of many unspecific protein interactions with chosen resin, BLUF-cI repressor was not purified successfully. However, after starting to use magnetic beads, we managed to obtain purified protein, reaching the concentration up to 0.3 mg ml-1.
SELEX
To determine if our protein of interest has the ability to bind DNA, SELEX experiments were performed. For this, single-stranded oligonucleotides containing a 30-nt random sequence and specific primers were chemically synthesized and used to amplify the double-stranded oligonucleotide library.
Many different conditions of SELEX, using magnetic beads or antibodies, were tested by error and trial method. It turned out that the most commonly used techniques are not suitable for our protein. Regarding this, during the experiments, blocking agents and buffers were altered. The changes resulted in elution of target oligonucleotides after exposing samples to blue light (Figure 3).

Figure 3. Gel electrophoresis after performing the first cycle of SELEX. W1-W12 – wash fractions collected in the dark; E–E3 – elution fractions collected after exposure to blue light (from separate wells).
EMSA
We tried to perform EMSA by using Gel Red fluorescent stain, yet no shift, compared to the control sample was detected (Figure 4).

Figure 4. Fluorescent EMSA, visualized using GelRed DNA stain.
To achieve a higher sensitivity of the assay, we aimed to perform EMSA with radiolabelled oligonucleotides. Here we received help from last year Vilnius-Lithuania iGEM team member Tomas Venclovas, who is familiar with this method and has a license to work with radioactive materials. The binding reactions were performed both in the light and dark, with serial protein dilutions (Figure 5).

Figure 5. Results of EMSA, carried out with radioactive-labelled oligonucleotides. A – binding reactions performed in the light; B – binding reactions performed in the dark. K – oligonucleotide samples (without BLUF-cI).
While analysing the results, no DNA shifting was detected. We believe that this method needs to be optimized.
References:
1. Pellegrini-Calace M, Thornton JM. Detecting DNA-binding helix–turn–helix structural motifs using sequence and structure information. Nucleic Acids Res. 2005;33(7):2129-2140. doi:10.1093/nar/gki349
2. Wintjens R, Rooman M. Structural Classification of HTH DNA-binding Domains and Protein – DNA Interaction Modes. Journal of Molecular Biology. 1996;262(2):294-313. doi:10.1006/jmbi.1996.0514
3. Domratcheva T, Grigorenko BL, Schlichting I, Nemukhin AV. Molecular Models Predict Light-Induced Glutamine Tautomerization in BLUF Photoreceptors. Biophysical Journal. 2008;94(10):3872-3879. doi:10.1529/biophysj.107.124172
4. Park S-Y, Tame JRH. Seeing the light with BLUF proteins. Biophys Rev. 2017;9(2):169-176. doi:10.1007/s12551-017-0258-6
5. Yee EF, Chandrasekaran S, Lin C, Crane BR. Physical methods for studying flavoprotein photoreceptors. Meth Enzymol. 2019;620:509-544. doi:10.1016/bs.mie.2019.03.023
6. Christie JM, Gawthorne J, Young G, Fraser NJ, Roe AJ. LOV to BLUF: Flavoprotein Contributions to the Optogenetic Toolkit. Molecular Plant. 2012;5(3):533-544. doi:10.1093/mp/sss020