Abstract
Diabetes Mellitus is one of the main diseases that causes premature death worldwide [1]. In 2014, worldwide 422 Million adults were diagnosed with Diabetes Mellitus [2], an alarmingly high number. This inspired us to look into alternative treatment strategies for Type 2 Diabetes Mellitus, as it marks the largest group of Diabetes patients. There already is a variety of treatment options, however, they all require active participation of the patients and compliance to their individual therapy schemes. As compliance and persistence are key factors for therapeutic success, we, the iGEM Team Tübingen, want to revolutionize the treatment and its administration with the use of Synthetic Biology. Therefore, we developed GLP.exe, a probiotic on the basis of Escherichia coli Nissle 1917, which secretes Exendin-4, an incretin analog, in response to glucose. To ensure its safe use as a GMO, we integrated a CRISPR/Cas3-based kill-switch into our probiotic. This kill-switch is regulated by environmental factors, and prevents the release and spread of our GMO into the environment. Our probiotic allows for a single-time application, as it synthesizes the drug independently when it is needed. To deepen our understanding of our probiotic strain, we also characterized E. coli Nissle 1917 by different means such as RNA-Seq and metabolic modeling. This characterization will be beneficial for the iGEM and general scientific community in future projects. The use of GMOs, especially in the medical field, is a delicate topic and we made great efforts during our project to increase awareness, and to educate the public about synthetic biology, GMOs, and Diabetes.
Project Inspiration
When committing to iGEM, we decided that we firstly wanted to design a project, which uses a novel concept in the field of synthetic biology, and secondly to make an impact with a product, which can actually have an impact for the society. Dr. Pengfei Xia, one of our advisors, proposed to design a new kill-switch system for bacteria, based on the Type I CRISPR system (CRISPR/Cas3). Immediately, we were determined to not only implement the system, but also find a way of making it a useful, universally applicable, tool. We decided to focus our project on Type 2 Diabetes Mellitus, because some relatives of our team members suffer from it, and this disease becomes increasingly epidemic, and a problem for society and the healthcare system.
We thought about changes in therapy, which would make it easier to comply with the therapy scheme. Eventually, we identified issues such as the required daily injections and multiple drugs, which need to be taken at certain times during the day. Overall, we came to the conclusion that a probiotic bacterium, which synthesizes a drug on demand, would release the burden of application from the patient. On top of that, we realized that in 2017 Team AQA_Unesp had already tried to target Type 1 Diabetes Mellitus with a probiotic. We felt that the viability of our idea was supported by this [3]. Importantly, this approach would allow us to use our bacterium with the CRISPR/Cas3 system as a biofactory, and simultaneously turn it into a therapeutic agent, which fulfills biosafety requirements for a safe application.
The idea to use GMOs as probiotics, is generally of great interest for chronic diseases. Therefore, we want to use the application in Type 2 Diabetes Mellitus as an example of the strengths and limitations of such a system. In combination with our public involvement, we want to gather the society’s perception and opinion of such a therapeutic strategy. Overall, we consider our project to be an important trial for the use of synthetic biology in long-term therapy during which the patient is not restricted to stay within a facility, but can live normally and without financial or other burdens of the disease. By designing a safe system, which will prohibit the survival of GMOs within the environment, our project aims at enabling the use of probiotics in modern therapy.
Glucose-dependent Incretin secretion
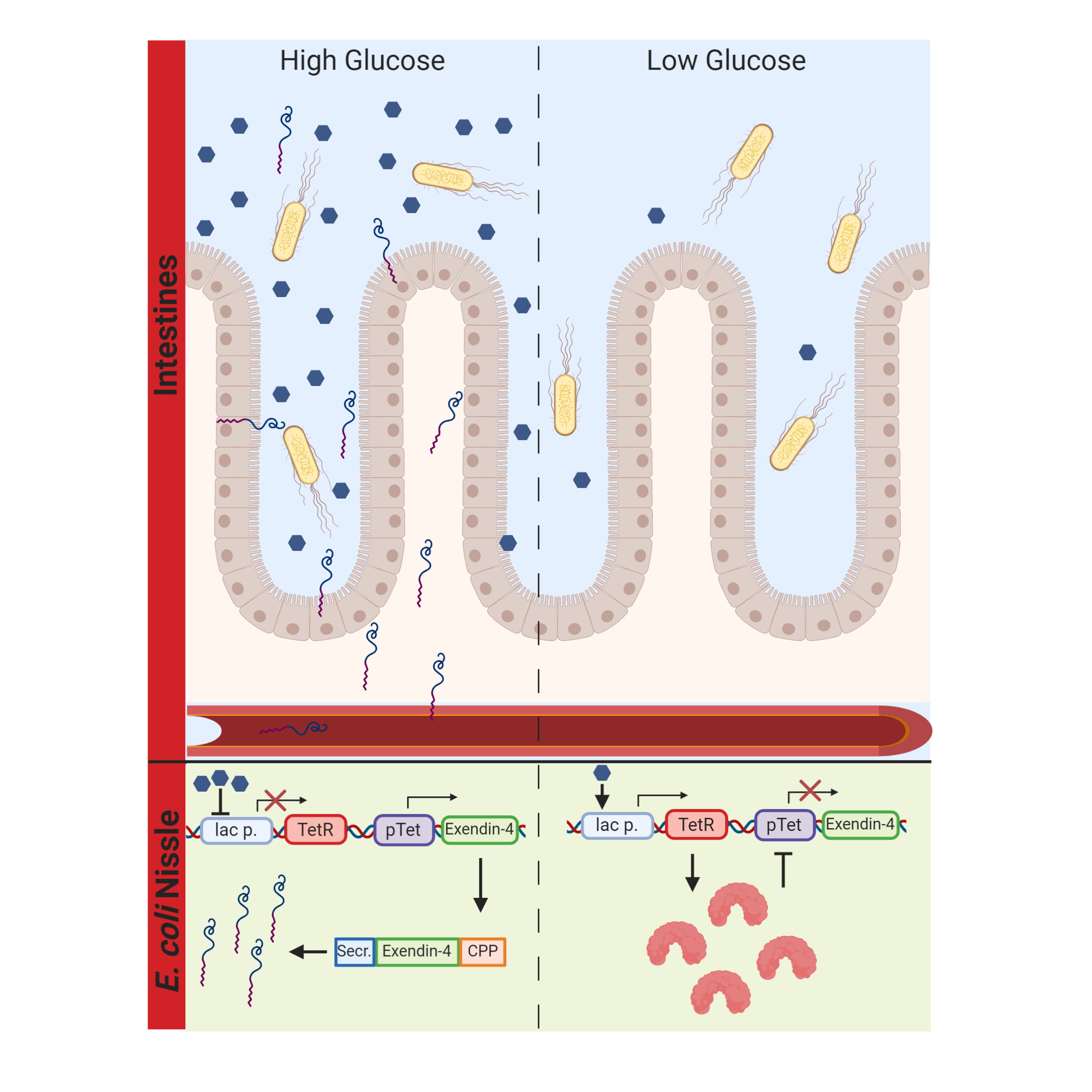
Our probiotic secretes Exendin-4, an Incretin mimetic, in response to glucose availability in the human gut. The system works via the carbon catabolite repression system, which will initiate the transcription of tetR. Upstream of our Exendin-4 is a TetR repressible promoter, which ensures that Exendin-4 is only transcribed in the presence of glucose. Furthermore, our Exendin-4 is coupled to an N-terminal secretion tag and a C-terminal cell penetrating peptide (CPP). The secretion tag ensures the secretion of Exendin-4 into the gut, while the CPP ensures the uptake of Exendin-4 into the bloodstream, which increases the overall bioavailability. Due to the immense variety of CPPs and the lack of quantitative information about their efficiency, we also developed a predictive software tool to allow for educated decisions on the design of CPPs.
CRISPR/Cas3 kill-switch
The implemented and newly developed CRISPR/Cas3 kill-switch allows for the safe use of our probiotic. Once the kill-switch is activated, the Cas3 nuclease degrades the bacterial DNA, and therefore prevents the spread of GMOs into the environment. The kill-switch is regulated by three environmental factors, which are common in a healthy humans intestine: 1) 37°C; 2) availability of fatty acids in form of Acyl-CoA; and 3) N-Acetyl-Glucosamin (GlcNAc), which is released by the metabolism of mucus through commensal microorganisms [9]. As soon as the probiotic leaves its designated area, the repression of the kill-switch is abrogated and the CRISPR/Cas3 system is activated. The kill switch, therefore, is a concept that can be used for a diverse spectrum of therapies by exchanging the drug and/or the conditions.

E. coli Nissle Characterisation
Despite the wide use of E. coli Nissle 1917 (EcN) it is not characterized well enough. To enhance our knowledge of EcN, and to provide crucial information to the scientific community as well as future iGEM teams, we investigated EcN’s transcriptome under different stress conditions. Understanding the complete transcriptome, the expressed genes, post-transcriptional modifications, single-nucleotide polymorphisms (SNPs), and additional properties of interest is essential for understanding genetic causes of adaptations to stress. The gained insight could lead to the development of more stress resistant strains, improving probiotic treatment to a large degree. Hence, we conducted large-scale RNA-Seq experiments for 12 conditions, including aerobic and anaerobic environments. The conditions were chosen after a thorough evaluation of the growth of EcN at different levels of the stress conditions. Additionally, we created the first ever metabolic model of EcN and made it available to the iGEM and general scientific community. Moreover, we modelled the interaction of EcN with three different bacterial communities - which is cutting-edge research and a novelty in the iGEM competition.
Software
To transport Exendin-4 across the membrane of the enterocytes in the gut, we decided to utilize cell-penetrating peptides (CPPs) as a carrier. CPPs have already been proven to transport different cargos, such as insulin, from the gut to the bloodstream [98]. To better understand the mechanism of action of CPPs and to make an educated decision for choosing the CPP domain in our project design, we decided to generate a machine learning based model to predict the cargo transport efficiency.
CPPs gain more and more attention in the scientific field, with multiple peptides being in clinical trials to deliver drug molecules to target sites in patients [99]. Since they can be used in various transport scenarios, many iGEM teams submitted CPPs to the registry in the past years. Therefore, we made our transport effectivity quantification software available to all future iGEM teams and the scientific community. To ensure excellent usability, we implemented a web GUI that allows multiple input formats.

Human Practices & Public Outreach
Because our project involves the use of GMOs, which are a constant topic of debate, we realized that we needed to invest into increasing awareness and especially education about GMOs, synthetic biology, and Diabetes. We therefore contacted several experts and teamed up with a variety of institutions, researchers and also iGEM teams in order to address this. Several collaborations, meetups, exchanges and talks provided us with valuable information for our project and helped developing it over the year. Learn more about our Human Practices under Human Practices / Overview.
Wetlab Project Plan
Our project required intensive cloning of multiple regulatory elements. For the parts, we used Biobricks that were sent with the 2019 shipping, Biobrick sequences from the database of iGEM, as well as new sequences, which are unique to our project, such as the Exendin-4 fusion construct or the GlcNAc-6-P sensing system. After synthesis, the GOI constructs were amplified and cloned into vectors. These were used to transform competent E. coli cells that are commonly used for cloning applications, as they can be manipulated easily. The design of all parts is finished and the glucose-dependent TetR expression as well as the Exendin-4 construct can be tested. Those two parts just need to be combined to one part to have the fully functional glucose-dependent Exendin-4 expression system. This system can then be tested for its glucose responsiveness and Exendin-4 expression as a next step.
At the same time the Cas3 system was isolated from E. coli K12, and the DNA with regulatory system was changed in E. coli K12 to our individual sensing mechanisms. All parts essentially needed for the CRISPR/Cas system have been designed and are also associated with their respective promoters. The regulatory circuit that controls the kill switch is fully designed but not yet tested since the ligation of the needed parts was not completed yet. The functionality of the kill switch and the different conditions for it can be tested when the regulatory circuit is cloned together. Individual testing of the separate regulators will be one of the next steps before combining all regulators and the CRISPR/Cas3 system into one system. If the system shows the desired activity, it will be integrated into the E. coli Nissle 1917 genome.
When all parts are tested individually, we hope to combine our GOI with the Cas3 positive E. coli Nissle 1917 and test the functionality of our system as a whole. More precisely, we want to test if the generated strain secretes Exendin-4 in the presence of glucose, and if it destructs itself under conditions diverting from the gut.
In the future, we will proceed with experiments on human cell lines. Firstly, the transport of Exendin-4 through a CaCo-2 cell monolayer will be tested to investigate whether this drug can reach the pancreas. Secondly, we will work with Rat Insulinoma cell line INS-1 cells to examine whether Exendin-4 will induce an Insulin expression.
References
- World Health Organization. (2016). Global Report on Diabetes. Available online; [Accessed 26.03.2019].
- Jens Juul Holst. The Physiology of Glucagon-like Peptide 1. (2007). Physiological Reviews. p.1409-1439.
- Lim, Gareth E., Brubaker, Patricia L. Glucagon-Like Peptide 1 Secretion by the L-Cell. (2006). 10.2337/db06-S020. Diabetes. p. S70-S77
- Copley, Kathrin & McCowen, Kevin & Hiles, Richard & L Nielsen, Loretta & Young, Andrew & Parkes, David. (2006). Investigation of Exendin-4e Elimination and Its In Vivo and In Vitro Degradation. Current drug metabolism. 7. 367-74. 10.2174/138920006776873490.
- Duan F, Curtis KL, March JC. Secretion of insulinotropic proteins by commensal bacteria: rewiring the gut to treat diabetes. Appl Environ Microbiol. (2008);74(23):7437–7438. doi:10.1128/AEM.01019-08
- Duan FF, Liu JH, March JC. Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes. (2015);64(5):1794–1803. doi:10.2337/db14-0635
- iGEM Team AQA_Unesp
- Team NTU-Taida
- Sicard JF, Le Bihan G, Vogeleer P, Jacques M, Harel J. Interactions of Intestinal Bacteria with Components of the Intestinal Mucus. Front Cell Infect Microbiol. (2017);7:387. Published 2017 Sep 5. doi:10.3389/fcimb.2017.00387
- Barnhart MM, Lynem J, Chapman MR. GlcNAc-6P levels modulate the expression of Curli fibers by Escherichia coli. J Bacteriol. (2006);188(14):5212–5219. doi:10.1128/JB.00234-06
- Konopka JB. N-acetylglucosamine (GlcNAc) functions in cell signaling. Scientifica (Cairo). (2012);2012:489208. doi:10.6064/2012/489208
- uniprot
- Feng Y, Cronan JE. Crosstalk of Escherichia coli FadR with global regulators in expression of fatty acid transport genes. PLoS One. (2012);7(9):e46275. doi:10.1371/journal.pone.0046275
- Federle MJ. Autoinducer-2-based chemical communication in bacteria: complexities of interspecies signaling. Contrib Microbiol. (2009);16:18–32. doi:10.1159/000219371
- Griffiths AJF, Miller JH, Suzuki DT, et al. An Introduction to Genetic Analysis. 7th edition. New York: W. H. Freeman; (2000). Catabolite repression of the lac operon: positive control. Available from:
- IDF (International Diabetes Federation. IDF Diabetes Atlas. (2017). Eighth Edition. Available from:
- Experimenta
- UN-SDG
- Tuebingen:Human_Practices
- Team CostaRica SDG
- Team Taipei video conference
- Jenabioscience































