| (33 intermediate revisions by 4 users not shown) | |||
| Line 3: | Line 3: | ||
<div> | <div> | ||
<div class="box-dark"> | <div class="box-dark"> | ||
| − | |||
<h1 class="heading"> | <h1 class="heading"> | ||
H U M A N   P R A C T I C E S | H U M A N   P R A C T I C E S | ||
</h1> | </h1> | ||
<hr class="line"> | <hr class="line"> | ||
| − | <img src="https://static.igem.org/mediawiki/2019/a/ac/T--Marburg--logo.svg" class="logo" alt="Syntex Logo"> | + | <img src="https://static.igem.org/mediawiki/2019/a/ac/T--Marburg--logo.svg" |
| + | class="logo" | ||
| + | alt="Syntex Logo"> | ||
</div> | </div> | ||
<div style="margin-top: 11vh;"> | <div style="margin-top: 11vh;"> | ||
<section class="section"> | <section class="section"> | ||
| − | + | <h1 class="title">Report on Genetic Engineering</h1> | |
| − | + | <p style="text-align: justify;"> | |
| − | + | Genetic engineering has been a hotly debated topic in politics as well as society in the past decades and | |
| − | + | still is today. | |
| − | + | Arguments like the nutrition of a growing world population due to a declining infant mortality rate or the | |
| − | + | loss of | |
| − | + | considerable areas of arable land due to erosion or pollution damage keep fueling the controversy whether | |
| − | + | genetically modified | |
| − | + | organisms (GMO), especially crops, are needed to sustain the global demand for food. On the opposite, | |
| − | + | concerns | |
| − | + | have been raised | |
| − | + | concerning the potential adverse effects on human health and environmental safety. Besides the facts, part | |
| − | + | of | |
| − | + | the public dispute | |
| − | + | is based around ethical questions and trust issues towards institutions and authorities. There have been | |
| − | + | studies and surveys | |
| − | + | carried out addressing many of these topics. Additionally a diverse cluster of organisations and the | |
| − | + | media | |
| − | + | is bombarding the | |
| − | + | public with contrary statements. This report tries to give an overview on mankind's relation towards modifying | |
| − | + | genetics, a brief | |
| − | + | summary of used methods, and gathers statements from scientists and authorities. It is meant as the | |
| − | + | motivational basis for this | |
| − | + | years Marburg iGEM team´s Public Engagement and Human Practice efforts.<br> | |
| − | + | </p> | |
| − | + | ||
| − | + | ||
</section> | </section> | ||
<hr> | <hr> | ||
<section class="section grid"> | <section class="section grid"> | ||
| − | <div class="sub" onclick="popup('gmo_report')"> | + | <div class="sub" |
| + | onclick="popup('gmo_report')"> | ||
<div class="sub-header"> | <div class="sub-header"> | ||
<h1> | <h1> | ||
| Line 52: | Line 52: | ||
<div class="sub-content"> | <div class="sub-content"> | ||
<div> | <div> | ||
| − | Gathering reviews, opinions and statements, this report is meant as the foundation of our Human Practice efforts. | + | Gathering reviews, opinions and statements, this report is meant as the foundation of our Human Practice |
| + | efforts. | ||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <div id="gmo_report" class="popup"> | + | <div id="gmo_report" |
| + | class="popup"> | ||
<div class="popup-container"> | <div class="popup-container"> | ||
<div class="popup-header"> | <div class="popup-header"> | ||
<h1 class="title">GMO Report</h1> | <h1 class="title">GMO Report</h1> | ||
| − | <button type="button" onclick="hide('gmo_report')">X</button> | + | <button type="button" |
| + | onclick="hide('gmo_report')">X</button> | ||
</div> | </div> | ||
| − | <div class="popup-content" style="text-align: justify;"> | + | <div class="popup-content" |
| + | style="text-align: justify;"> | ||
<section class="section"> | <section class="section"> | ||
| − | <div> | + | <div> |
| − | + | <p style="text-align: justify; margin-bottom: 1em;"> | |
| − | + | <b>History of Genetic Modification</b><br> | |
| − | + | Our ancestors had no conception of genetics but still were able to influence the genes of multiple | |
| − | + | organisms. | |
| − | + | It is a | |
| − | + | process known to everybody called artificial selection or selective breeding. Those individuals with | |
| − | + | the | |
| − | + | most | |
| − | + | desirable traits, | |
| − | + | like the biggest and most delicious fruits or the highest loyalty, is chosen to propagate and | |
| − | + | produce | |
| − | + | offspring. This process | |
| − | + | is repeated over several generations and the result is an organism with the selected traits. The | |
| − | + | dog, | |
| − | + | existing | |
| − | + | today in many | |
| − | + | variations, is believed to be the organism our ancestors selectively bred first around 32,000 years | |
| − | + | ago <a style="padding: 0" | |
| − | + | href="https://www.nytimes.com/2013/05/16/science/dogs-from-fearsome-predator-to-mans-best-friend.html" | |
| − | + | target="_blank">(Zimmer, 2013)</a>. And there are many more instances like corn which originates | |
| − | + | from a grass called teosinte | |
| − | + | with | |
| − | + | very few kernels <a style="padding: 0" | |
| − | + | href="https://learn.genetics.utah.edu/content/evolution/corn/" | |
| − | + | target="_blank">(‘Evolution of | |
| − | + | Corn’, | |
| − | + | n.d.)</a>. However, this process is not considered GMO technology today. What we understand under | |
| − | + | genetic | |
| − | + | modification today can be traced back to | |
| − | + | the mid 1900´s, | |
| − | + | when scientists discovered that genetic material can be transferred between different species | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1084/jem.79.2.137" | |
| − | + | target="_blank">(Avery, MacLeod, & McCarty, 1944)</a>, | |
| − | + | the structure of genetic material was identified as a double helix <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1098/rspa.1954.0101" | |
| − | + | target="_blank">(Crick, Watson, & Bragg, 1954)</a>, the genetic code was | |
| − | + | deciphered <a style="padding: 0" | |
| − | + | href="https://www.ncbi.nlm.nih.gov/pubmed/13938750" | |
| − | + | target="_blank">(Nirenberg, Matthaei, | |
| − | + | Jones, Martin, & | |
| − | + | Barondes, 1963)</a> and finally a DNA recombinant technology was described <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1073/pnas.70.11.3240" | |
| − | + | target="_blank">(Cohen, Chang, Boyer, & Helling, 1973)</a>. Only a few | |
| − | + | decades after these | |
| − | + | ground-breaking discoveries were made, the first | |
| − | + | genetically modified (GM) plants were produced in 1983, which were antibiotic resistant tobacco and | |
| − | + | petunia | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://www.ncbi.nlm.nih.gov/pubmed/7153688" | |
| − | + | target="_blank">(Bevan & Chilton, 1982; Fraley, | |
| − | + | 1983; Herrera‐Estrella | |
| − | + | et al., 1983)</a>. | |
| − | + | Soon, the first GM plants were commercialized: In the | |
| − | + | early 1990´s China approved modified tobacco and in 1994 the United States Food and Drug | |
| − | + | Administration | |
| − | + | (U.S. | |
| − | + | FDA) approved | |
| − | + | the “FLAVR SAVR” tomato which was modified to have a longer shelf live by delaying ripening. Today, | |
| − | + | numerous | |
| − | + | GM | |
| − | + | plants exist | |
| − | + | and are in use, covering popular fruits like papaya, melon and apple, flowers like roses, feed | |
| − | + | plants like | |
| − | + | sugar beet, | |
| − | + | vegetables like tomato, maize and potato and even cotton for clothes production | |
| − | + | <a style="padding: 0" | |
| − | + | href="http://www.isaaa.org/gmapprovaldatabase/cropslist/default.asp" | |
| − | + | target="_blank">(‘GM Crops | |
| − | + | List—GM Approval | |
| − | + | Database | ISAAA.org’, n.d.)</a>.</p> | |
| − | + | <p style="text-align: justify; margin-bottom: 1em;"> | |
| − | + | <b>Current Numbers on GM Crops</b><br> | |
| − | + | <i>World</i><br>As stated above, many GM crops are relevant for food production today, be it | |
| − | + | indirectly | |
| − | + | for | |
| − | + | animal feed in | |
| − | + | production lines or directly as consumables. In 2018, 26 countries planted 191.7 million hectares | |
| − | + | worldwide | |
| − | + | with GM crops, | |
| − | + | which is an increase of 1% from 2017´s worldwide planted area. Accordingly, since its first | |
| − | + | commercialization | |
| − | + | in 1996 with | |
| − | + | 1.7 million hectares planted, GM crop area increased by an approximate 113-fold. The accumulated | |
| − | + | area | |
| − | + | planted | |
| − | + | with GM crops | |
| − | + | from 1996 to 2018 was 2.5 billion hectares. This makes biotechnology the fastest adopted crop | |
| − | + | technology | |
| − | + | in | |
| − | + | the world. Of | |
| − | + | the 193 member nations of the United Nations Organisation (UNO) 42 nations plus the European Union | |
| − | + | (EU) | |
| − | + | adopted GM crops, | |
| − | + | of which 26 countries (21 developing and 5 industrial) planted and 44 imported GM crops. The four | |
| − | + | major GM | |
| − | + | crops, namely | |
| − | + | soybeans, maize, cotton and canola, occupied 99% of the GM crop area (Figure 1). GM crop share in | |
| − | + | total | |
| − | + | crop | |
| − | + | area was 78% | |
| − | + | for soybeans, 76% for cotton, 30% for maize and 29% for canola. 42% of the global GM crop area was | |
| − | + | planted | |
| − | + | with | |
| − | + | stacked trait | |
| − | + | crops tolerant to various herbicides and pesticides. Around the world the GM crop area was unevenly | |
| − | + | distributed with the top | |
| − | + | five countries United States of America (USA), Brazil, Argentina, Canada and India planting 91% of | |
| − | + | the | |
| − | + | global | |
| − | + | GM crop area. | |
| − | + | In the EU, the two nations Spain and Portugal planted the GM crop MON810, which is an insecticide | |
| − | + | resistant | |
| − | + | maize, together | |
| − | + | covering 120.990 hectares. 95% of the area was planted by Spain. From 2017 to 2018 GM crop area in | |
| − | + | the EU | |
| − | + | has | |
| − | + | decreased by | |
| − | + | 8% from 131.535 hectares (Figure 2). Nevertheless the EU imported GM crops, roughly 30 million tons | |
| − | + | of | |
| − | + | soybean | |
| − | + | products, | |
| − | + | 10 million tons of maize and 2.5 million tons of canola originating from Argentina, Brazil and the | |
| − | + | USA. | |
| − | + | Since | |
| − | + | 1992, across | |
| − | + | the world 4.349 approvals to GM crops have been issued, of this being 2.063 for food, 1.461 for | |
| − | + | animal feed use | |
| − | + | and | |
| − | + | 825 for | |
| − | + | cultivation <a style="padding: 0" | |
| − | + | href="https://www.isaaa.org/resources/publications/briefs/54/executivesummary/default.asp" | |
| − | + | target="_blank"> | |
| − | + | (‘ISAAA Brief 54-2018: Executive Summary | ISAAA.org’, n.d.)</a>. | |
| − | + | </p> | |
| − | + | <figure style="text-align:center"> | |
| − | + | <img style="height: 393px; width: 450px;" | |
| − | + | src="https://static.igem.org/mediawiki/2019/b/b3/T--marburg--report_on_genetic_engineering_GM_crops_2018.jpg" | |
| − | + | alt="GM crops 2018"> | |
| − | + | <figcaption style="max-width: 2400px; text-align: center"> | |
| − | + | Fig.1 - Area and adoption rate of GM crops (biotech crops) in 2018 worldwide. | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://www.isaaa.org/resources/publications/briefs/54/executivesummary/default.asp" | |
| − | + | target="_blank"> | |
| − | + | Adopted from ISAAA, 2018</a>. <br>*GM sugar beets, potatoes, | |
| − | + | apple, squash, papaya and brinjal/eggplant. | |
| − | + | </figcaption> | |
| − | + | </figure> | |
| − | + | <figure style="text-align:center"> | |
| − | + | <img style="height: 500px; width: 500px;" | |
| − | + | src="https://static.igem.org/mediawiki/2019/0/00/T--marburg--report_on_genetic_engineering_global_map.jpg" | |
| − | + | alt="Global map"> | |
| − | + | <figcaption style="max-width: 2400px; text-align: center"> | |
| − | + | Fig.2 - Global map of GM (biotech) countries in 2018. | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://www.isaaa.org/resources/publications/briefs/54/executivesummary/default.asp" | |
| − | + | target="_blank"> | |
| − | + | Adopted from ISAAA, 2018</a>. | |
| − | + | </figcaption> | |
| − | + | </figure> | |
| − | + | <p style="text-align: justify; margin-bottom: 1em;"> | |
| − | + | <i>Germany</i><br> | |
| − | + | In Germany, there is no more GM crop farming since 2012. GM maize has been planted last in 2008 | |
| − | + | (3.171 hectares, 0.15% of total maize area in Germany) and GM potatoes have been planted last in | |
| − | + | 2011 | |
| − | + | (2 hectares, 0.0008% of total potato area in Germany). GM crop area never made up more than 0.02% of | |
| − | + | land | |
| − | + | used | |
| − | + | by | |
| − | + | agriculture in Germany <a style="padding: 0" | |
| − | + | href="https://www.bmel.de/DE/Landwirtschaft/Pflanzenbau/Gentechnik/_Texte/Gentechnik_Wasgenauistdas.html" | |
| − | + | target="_blank"> | |
| − | + | (‘Gentechnik’, n.d.)</a>.</p> | |
| − | + | <p style="text-align: justify; margin-bottom: 1em;"> | |
| − | + | <b>Modern Methods in Breeding</b><br> | |
| − | + | The traditional way of breeding, as explained above, although having generated many domestic plants | |
| − | + | and animals, | |
| − | + | is | |
| − | + | relatively | |
| − | + | slow and limited by the available traits individuals express. Modern breeding methods enhance the | |
| − | + | trait | |
| − | + | spectrum and the pace | |
| − | + | in which new traits can be discovered and implemented to crops and animals. </p> | |
| − | + | <p style="text-align: justify; margin-bottom: 1em;"> | |
| − | + | <i>Plant Mutagenesis</i><br> | |
| − | + | As it is known that practical breeding depends on genetic variation plant mutagenesis expands the | |
| − | + | variability of | |
| − | + | traits. | |
| − | + | Variations found in nature do not represent the original spectra of spontaneous mutations due to the | |
| − | + | fact | |
| − | + | that | |
| − | + | they are | |
| − | + | recombining within populations and interacting with environmental factors. In the process of | |
| − | + | mutagenesis | |
| − | + | heritable changes | |
| − | + | occur in the genetic information induced by mutagenic agents called mutagens. These mutagens can be | |
| − | + | of | |
| − | + | chemical, | |
| − | + | for | |
| − | + | instance substances interacting with the DNA, or of physical origin, such as ionizing radiation | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1080/13102818.2015.1087333" | |
| − | + | target="_blank">(Oladosu et al., 2016)</a>. | |
| − | + | After using the mutagen on the crops, mostly seeds, seedlings or cell cultures from which single | |
| − | + | cells can | |
| − | + | be | |
| − | + | grown out, | |
| − | + | screening has to be done to see if changes in traits have been achieved by mutations. These | |
| − | + | mutations can | |
| − | + | be | |
| − | + | DNA | |
| − | + | double | |
| − | + | strand breaks, single base exchanges or alkylation of bases. In most cases, generated mutants are | |
| − | + | heterozygous, | |
| − | + | because | |
| − | + | the mutation happened in only one allele. Therefore the breeder needs to rear subsequent generations | |
| − | + | to | |
| − | + | evaluate | |
| − | + | recessive | |
| − | + | mutations. Selection then takes place in form of phenotypical, physical or molecular testing to | |
| − | + | determine for | |
| − | + | instance plant | |
| − | + | height, earliness of maturity and biochemical composition. Mutagenesis breeding has impacted | |
| − | + | agriculture | |
| − | + | massively with | |
| − | + | more than 3.300 entries to the Mutant Variety Database | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://mvd.iaea.org/#!Search?page=1&size=500&sortby=Name&sort=ASC" | |
| − | + | target="_blank">(‘Mutant | |
| − | + | Variety Database’, | |
| − | + | n.d.)</a>, | |
| − | + | covering all major food and feed crops.</p> | |
| − | + | <p style="text-align: justify; margin-bottom: 1em;"> | |
| − | + | <i>Genetic Engineering</i><br> | |
| − | + | This term is used to describe methods which alter the genetic makeup of an organism using DNA | |
| − | + | recombinant | |
| − | + | technology. | |
| − | + | This technology resorts to enzymatic tools called restriction enzymes. These cut the DNA site | |
| − | + | specific and | |
| − | + | can | |
| − | + | thereby | |
| − | + | isolate genetic constructs coding for desirable traits. When gene(s) are introduced into an organism | |
| − | + | this | |
| − | + | can be | |
| − | + | achieved | |
| − | + | either directly or indirectly. The direct approach utilizes a method called microparticle | |
| − | + | bombardment | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1111/j.1399-3054.1990.tb05888.x" | |
| − | + | target="_blank">(Sanford, 1990)</a>. | |
| − | + | Developed in the 1980´s, engineered DNA is coated on microparticles of either gold or tungsten and | |
| − | + | then | |
| − | + | shot with high velocity at the target organism using high pressure helium gas. The DNA fragments can | |
| − | + | then | |
| − | + | be | |
| − | + | incorporated | |
| − | + | into the organism’s genetic material. There are other direct methods such as electroporation or | |
| − | + | microinjection | |
| − | + | but particle | |
| − | + | bombardment is the most effective. The indirect approach makes use of a vector: the soil bacterium | |
| − | + | <i>Agrobacterium | |
| − | + | tumefaciens</i> | |
| − | + | naturally infects plants and alters its hosts genome via a plasmid called Ti-plasmid. This plasmid | |
| − | + | can be | |
| − | + | engineered to carry | |
| − | + | genes coding for a desired trait instead of its natural genes for infection. With the development of | |
| − | + | a | |
| − | + | method | |
| − | + | called | |
| − | + | CRISPR/Cas9 and other variants genetic engineering in plants got much easier | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1126/science.1231143" | |
| − | + | target="_blank">(Cong et al., 2013</a>; | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1095/biolreprod.114.123935" | |
| − | + | target="_blank">DeMayo & Spencer, 2014</a>; | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1038/nprot.2013.143" | |
| − | + | target="_blank">Ran et al., 2013)</a>. | |
| − | + | This system is found in bacteria where it serves as a defence mechanism against viruses. The | |
| − | + | endonuclease | |
| − | + | is | |
| − | + | guided to its | |
| − | + | target cutting site via a guide RNA where it induces a double strand break (DBS). The DBS can be | |
| − | + | repaired | |
| − | + | in | |
| − | + | two distinct | |
| − | + | ways. Non-homologous end joining leads to a small deletion while homologous recombination allows for | |
| − | + | the | |
| − | + | integration of | |
| − | + | donor DNA into the endogenous DNA. Thereby, the CRISPR method allows for small alteration or whole | |
| − | + | gene | |
| − | + | insertions at target | |
| − | + | sites.<br>At this point it may be appropriate to introduce the two terms “cisgenic” and | |
| − | + | “transgenic”. | |
| − | + | While | |
| − | + | “transgenic” | |
| − | + | refers to organisms in which genetic material outside the species boundary, originating from a donor | |
| − | + | organism | |
| − | + | which is | |
| − | + | sexually incompatible to the engineered organism, has been inserted. “Cisgenic” on the contrary | |
| − | + | describes | |
| − | + | genetic | |
| − | + | modifications within the boundaries of sexual compatibility. Therefore, cisgenic plants are similar | |
| − | + | to | |
| − | + | traditionally bred | |
| − | + | plants <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1038/sj.embor.7400769" | |
| − | + | target="_blank">(Schouten, Krens, & Jacobsen, | |
| − | + | 2006)</a>. The most | |
| − | + | obvious example | |
| − | + | of transgenic plants are the many varieties of so | |
| − | + | called “Bt” crops. Standing for <i>Bacillus thuringiensis</i>, into these plants a gene from the | |
| − | + | bacterium was | |
| − | + | integrated which | |
| − | + | leads to the production of a crystal protein that is toxic to specific pest insects | |
| − | + | <a style="padding: 0" | |
| − | + | href="http://sitn.hms.harvard.edu/flash/2015/insecticidal-plants/" | |
| − | + | target="_blank">(‘Insecticidal | |
| − | + | Plants’, 2015)</a>.</p> | |
| − | + | <p style="text-align: justify; margin-bottom: 1em;"> | |
| − | + | <b>Opinions on GMOs</b><br> | |
| − | + | There are many scientific publications evaluating specific GMO traits towards the environment and | |
| − | + | health | |
| − | + | safety. | |
| − | + | Additionally many reviews exist summarizing GMO effects to a much broader scale possible here | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1007/s13197-012-0899-1" | |
| − | + | target="_blank">(Bawa & Anilakumar, 2013</a>; | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.3109/07388551.2013.823595" | |
| − | + | target="_blank">Nicolia, Manzo, Veronesi, & | |
| − | + | Rosellini, 2014</a>; | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1016/j.fct.2011.11.048" | |
| − | + | target="_blank">Snell et al., 2012</a>; | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1016/j.fshw.2016.04.002" | |
| − | + | target="_blank">Zhang, Wohlhueter, & Zhang, | |
| − | + | 2016)</a>. | |
| − | + | In many of these, authors conclude that the application of GMOs offers great opportunities but still | |
| − | + | has to | |
| − | + | be | |
| − | + | carried out | |
| − | + | with precautions. A simple “yes” or “no” cannot be given | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1016/j.fshw.2016.04.002" | |
| − | + | target="_blank">(Zhang et al., 2016)</a>. Still, | |
| − | + | due | |
| − | + | to the partly | |
| − | + | contradictory | |
| − | + | evidence, it cannot be said there is a consensus among scientists, according to | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1186/s12302-014-0034-1" | |
| − | + | target="_blank">Hilbeck et al., 2015</a>.</p> | |
| − | + | <p style="text-align: justify; margin-bottom: 1em;"> | |
| − | + | <i>Benefits of GM Crops</i><br> | |
| − | + | Humanity faces several challenges in the coming decades. Amongst them are the increasing world | |
| − | + | population, a | |
| − | + | decrease of | |
| − | + | arable land or the bottleneck of traditional breeding methods | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1016/j.fshw.2016.04.002" | |
| − | + | target="_blank">(Zhang et al., 2016)</a>. | |
| − | + | To all of these, GMOs pose a genuine answer. The easiest way to produce more food for a growing | |
| − | + | population | |
| − | + | is to | |
| − | + | increase | |
| − | + | productivity by earlier maturity, easier harvesting, processing and cultivation. Adding to that, if | |
| − | + | we | |
| − | + | resorted | |
| − | + | to | |
| − | + | organically producing todays yields, humanity would need to cultivate an additionally 3 billion | |
| − | + | hectares, | |
| − | + | which | |
| − | + | is the | |
| − | + | equivalent to the size of two South Americas | |
| − | + | <a style="padding: 0" | |
| − | + | href="http://www.marklynas.org/2013/04/time-to-call-out-the-anti-gmo-conspiracy-theory/" | |
| − | + | target="_blank">(‘Time | |
| − | + | to call | |
| − | + | out | |
| − | + | the anti-GMO conspiracy theory – Mark Lynas’, n.d.)</a>. | |
| − | + | But food also needs to become more nutritious. A good example here is “Golden Rice” | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1126/science.287.5451.303" | |
| − | + | target="_blank">(Ye et al., 2000)</a>, | |
| − | + | which produces a precursor of vitamin A. The deficiency of vitamin A is estimated to kill more than | |
| − | + | half a | |
| − | + | million | |
| − | + | children under the age of 5 each year | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1016/S0140-6736(07)61690-0" | |
| − | + | target="_blank">(Black et al., 2008)</a> | |
| − | + | and cause another half a million irreversible cases of childhood blindness | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://www.ncbi.nlm.nih.gov/pubmed/1600583" | |
| − | + | target="_blank">(Humphrey, West, & Sommer, | |
| − | + | 1992)</a>.</p> | |
| − | + | <p style="text-align: justify; margin-bottom: 1em;"> | |
| − | + | <i>Risks of GM Crops</i><br> | |
| − | + | GMOs pose risks to its consumer as do crops deriving from traditional breeding. Major risks are | |
| − | + | toxicity, | |
| − | + | allergenicity and genetic hazards emerging from the inserted or altered gene itself, the expressed | |
| − | + | protein, | |
| − | + | products | |
| − | + | of the metabolism, pleiotropic effects or the disruption of natural genes in the organism | |
| − | + | <a style="padding: 0" | |
| − | + | href="https://doi.org/10.1016/j.fshw.2016.04.002" | |
| + | target="_blank">(Zhang et al., 2016)</a>. | ||
| + | There have been reports on the strong allergenicity of “Starlink” maize, which is directly connected | ||
| + | to | ||
| + | the | ||
| + | inserted gene | ||
| + | from <i>Bacillus thuringiensis</i> | ||
| + | <a style="padding: 0" | ||
| + | href="https://doi.org/10.1016/j.toxicon.2006.11.022" | ||
| + | target="_blank">(Bravo, Gill, & Soberón, | ||
| + | 2007</a>; | ||
| + | <a style="padding: 0" | ||
| + | href="https://doi.org/10.1051/agro/2010027" | ||
| + | target="_blank">Sanchis, 2011</a>; | ||
| + | <a style="padding: 0" | ||
| + | href="https://doi.org/10.1146/annurev.en.39.010194.000403" | ||
| + | target="_blank">Tabashnik, 1994</a>; | ||
| + | <a style="padding: 0" | ||
| + | href="https://doi.org/10.1071/CP13167" | ||
| + | target="_blank">Werth, Boucher, Thornby, Walker, & Charles, | ||
| + | 2013)</a>. | ||
| + | Also, GM crops can have an adverse ecological influence. For example, the weed species <i>Amaranthus | ||
| + | palmeri</i> | ||
| + | did | ||
| + | evolve a | ||
| + | glyphosate resistance after years of glyphosate use on resistant cotton fields | ||
| + | <a style="padding: 0" | ||
| + | href="https://doi.org/10.1038/497024a" | ||
| + | target="_blank">(Gilbert, 2013)</a>. | ||
| + | Another possibility is the fact, that insect resistant crops infer with ecological food webs by | ||
| + | shifting | ||
| + | predator prey | ||
| + | ratios. Moreover, targeted pests might decline and primary minor pests become major issues | ||
| + | <a style="padding: 0" | ||
| + | href="https://doi.org/10.1007/s13197-012-0899-1" | ||
| + | target="_blank">(Bawa & Anilakumar, 2013; Snow & | ||
| + | Palma, 1997)</a>.</p> | ||
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| + | <b>Statements from Authorities</b><br> | ||
| + | The "Public Acceptance of Agricultural Biotechnologies (PABE) project" revealed a range of questions | ||
| + | concerning | ||
| + | rather institutional considerations of the public, such as who is benifitting from GMO use, by whom | ||
| + | consequences | ||
| + | have | ||
| + | been evaluated, if authorities have enough power to regulate large companies and why the public has | ||
| + | not | ||
| + | been | ||
| + | better | ||
| + | informed about GMO usage | ||
| + | <a style="padding: 0" | ||
| + | href="https://doi.org/10.1093/embo-reports/kve142" | ||
| + | target="_blank">(Marris, 2001)</a>. | ||
| + | For this reason, an overview of institutional statements might be appropriate.<br> | ||
| + | The European Commision (EC) published the book “A decade of EU-funded GMO research”. Within this | ||
| + | endeavor | ||
| + | more | ||
| + | than | ||
| + | 200 million Euro of research grants were spent to evaluate GMO´s in areas such as environmental | ||
| + | impact, | ||
| + | food | ||
| + | safety, | ||
| + | biomaterials, biofuels, risk assessment and management. It conclusively states: “The main conclusion | ||
| + | to be | ||
| + | drawn from the efforts of more than 130 research projects, covering a period of more than 25 years | ||
| + | of | ||
| + | research, | ||
| + | and | ||
| + | involving more than 500 independent research groups, is that biotechnology, and in particular GMOs, | ||
| + | are | ||
| + | not | ||
| + | per | ||
| + | se more | ||
| + | risky than e.g. conventional plant breeding technologies.” | ||
| + | <a style="padding: 0" | ||
| + | href="https://op.europa.eu:443/en/publication-detail/-/publication/d1be9ff9-f3fa-4f3c-86a5-beb0882e0e65" | ||
| + | target="_blank">(Publications | ||
| + | Office of the European Union, 2010)</a>.<br> | ||
| + | The National Academy of Sciences founded by the U.S. Congress summarizes in their comprehensive | ||
| + | report, | ||
| + | that | ||
| + | large | ||
| + | numbers of animal feeding studies provided reasonable evidence that animals were not harmed by food | ||
| + | derived | ||
| + | from | ||
| + | GM crops, | ||
| + | although admitting some studies were not designed optimal. Furthermore, long-term data in livestock | ||
| + | health | ||
| + | before and | ||
| + | after GM crop introduction did not show adverse effects associated with the crops. And at last, | ||
| + | epidemiological | ||
| + | data on | ||
| + | cancer and human health over time was revised but no substantiated evidence was found that GM crops | ||
| + | are | ||
| + | less | ||
| + | safe than | ||
| + | foods from non-GM crops | ||
| + | <a style="padding: 0" | ||
| + | href="https://doi.org/10.17226/23395" | ||
| + | target="_blank">(Read "Genetically Engineered Crops, | ||
| + | n.d.)</a>.<br> | ||
| + | The British Royal Society states the following to the question “Is it safe to eat GM crops?” on its | ||
| + | website: | ||
| + | “Yes. | ||
| + | There is no evidence that a crop is dangerous to eat just because it is GM. There could be risks | ||
| + | associated | ||
| + | with | ||
| + | the | ||
| + | specific new gene introduced, which is why each crop with a new characteristic introduced by GM is | ||
| + | subject | ||
| + | to | ||
| + | close scrutiny. | ||
| + | Since the first widespread commercialisation of GM produce 18 years ago there has been no evidence | ||
| + | of ill | ||
| + | effects linked to | ||
| + | the consumption of any approved GM crop.” Before new GM foods are permitted to the market a variety | ||
| + | of | ||
| + | test | ||
| + | has | ||
| + | to be | ||
| + | completed and the results are used by the authorities to determine the safety of the GM product, | ||
| + | making | ||
| + | “new | ||
| + | GM | ||
| + | crop | ||
| + | varieties at least as safe to eat as new non GM varieties, which are not tested in this way.” | ||
| + | <a style="padding: 0" | ||
| + | href="https://royalsociety.org/topics-policy/projects/gm-plants/is-it-safe-to-eat-gm-crops/" | ||
| + | target="_blank">(‘Is it | ||
| + | safe | ||
| + | to | ||
| + | eat GM crops?’, n.d.)</a>. | ||
| + | </p> | ||
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| + | <b>Conclusion</b><br> | ||
| + | As biologists, using genetic engineering methods every single day, they are quite natural to us. | ||
| + | Nevertheless, | ||
| + | we are | ||
| + | confronted with the public debate too. Having experienced the public aversion towards GMO ourselves | ||
| + | and | ||
| + | having | ||
| + | read about | ||
| + | the many proposed justifications against it we realized that a direct exchange between the public | ||
| + | and | ||
| + | experts | ||
| + | from all | ||
| + | fields as well as diverse interest groups might provide a good common ground for an open discussion. | ||
| + | <br> | ||
| + | </p> | ||
| + | </div> | ||
</section> | </section> | ||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <div class="sub" onclick="popup('nina_scheer')"> | + | <div class="sub" |
| + | onclick="popup('nina_scheer')"> | ||
<div class="sub-header"> | <div class="sub-header"> | ||
<h1> | <h1> | ||
| Line 492: | Line 645: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <div id="nina_scheer" class="popup"> | + | <div id="nina_scheer" |
| + | class="popup"> | ||
<div class="popup-container"> | <div class="popup-container"> | ||
<div class="popup-header"> | <div class="popup-header"> | ||
<h1 class="title">Dr. Nina Scheer</h1> | <h1 class="title">Dr. Nina Scheer</h1> | ||
| − | <button type="button" onclick="hide('nina_scheer')">X</button> | + | <button type="button" |
| + | onclick="hide('nina_scheer')">X</button> | ||
</div> | </div> | ||
| − | <div class="popup-content" style="text-align: justify;"> | + | <div class="popup-content" |
| + | style="text-align: justify;"> | ||
<section class="section"> | <section class="section"> | ||
<p style="text-align: justify; margin-bottom: 1em;"> | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| Line 512: | Line 668: | ||
preventing any further development and leaving important key technologies to global competitors.<br> | preventing any further development and leaving important key technologies to global competitors.<br> | ||
<br> | <br> | ||
| − | As we discussed with Dr. Scheer about our project and green genetical engineering in general, she supported the idea that | + | As we discussed with Dr. Scheer about our project and green genetical engineering in general, she |
| + | supported the idea that | ||
the state should invest more funding into research, rather than us relying on third-party funds. | the state should invest more funding into research, rather than us relying on third-party funds. | ||
| − | Nonetheless, Dr. Scheer did see a trust issue concerning irreversible damages to nature and humankind as | + | Nonetheless, Dr. Scheer did see a trust issue concerning irreversible damages to nature and humankind |
| + | as | ||
well as a lack of good control mechanisms when using green genetic engineering. At the example of | well as a lack of good control mechanisms when using green genetic engineering. At the example of | ||
| − | Contergan® Nina Scheer highlighted the challenges of uncertainty. We learned that we as scientists have | + | Contergan® Nina Scheer highlighted the challenges of uncertainty. We learned that we as scientists |
| + | have | ||
the duty to proof the unmitigated safety of our products and beyond that not only proof it to our | the duty to proof the unmitigated safety of our products and beyond that not only proof it to our | ||
own community but also to bring this trust to society as a whole if we want our research to have an | own community but also to bring this trust to society as a whole if we want our research to have an | ||
| Line 527: | Line 686: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <div class="sub" onclick="popup('plant_market')"> | + | <div class="sub" |
| + | onclick="popup('plant_market')"> | ||
<div class="sub-header"> | <div class="sub-header"> | ||
<h1> | <h1> | ||
| Line 536: | Line 696: | ||
<div class="sub-content"> | <div class="sub-content"> | ||
<div> | <div> | ||
| − | Reaching out to senior people, we advertised our panel discussion and adressed public concerns towards genetically modified organims. | + | Reaching out to senior people, we advertised our panel discussion and adressed public concerns towards |
| + | genetically modified organims. | ||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <div id="plant_market" class="popup"> | + | <div id="plant_market" |
| + | class="popup"> | ||
<div class="popup-container"> | <div class="popup-container"> | ||
<div class="popup-header"> | <div class="popup-header"> | ||
<h1 class="title">Plant Market</h1> | <h1 class="title">Plant Market</h1> | ||
| − | <button type="button" onclick="hide('plant_market')">X</button> | + | <button type="button" |
| + | onclick="hide('plant_market')">X</button> | ||
</div> | </div> | ||
| − | <div class="popup-content" style="text-align: justify;"> | + | <div class="popup-content" |
| − | < | + | style="text-align: justify;"> |
| − | <figure style="float: left; margin-right: 25px;"> | + | <section class="section"> |
| − | <img style=" | + | <figure style="float: left; margin-right: 25px;"> |
| + | <img style="width: 500px;" | ||
src="https://static.igem.org/mediawiki/2019/d/df/T--Marburg--plant_breeder_festival.jpeg" | src="https://static.igem.org/mediawiki/2019/d/df/T--Marburg--plant_breeder_festival.jpeg" | ||
alt="Extracting pepper"> | alt="Extracting pepper"> | ||
| − | <figcaption style="max-width: | + | <figcaption style="max-width: 500px;"> |
Fig.1 - Thomas Södler extracting DNA from pepper at the plant breeder festival. | Fig.1 - Thomas Södler extracting DNA from pepper at the plant breeder festival. | ||
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
<p style="text-align: justify; margin-bottom: 1em;"> | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| − | |||
We advertised our panel discussion by running a booth at the plant breeders festival in Marburg on the | We advertised our panel discussion by running a booth at the plant breeders festival in Marburg on the | ||
weekend of 14th September. It is an event for young and old people alike and the perfect platform to | weekend of 14th September. It is an event for young and old people alike and the perfect platform to | ||
| − | bring our discussion closer to a diverse audience.< | + | bring our discussion closer to a diverse audience. |
| − | + | </p> | |
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
When we talked with Nina Scheer, member of the German parliament from the Social Democratic Party, she | When we talked with Nina Scheer, member of the German parliament from the Social Democratic Party, she | ||
told | told | ||
| Line 573: | Line 737: | ||
know enough about it and would like to hear more. Through personal discussions we were able to get in | know enough about it and would like to hear more. Through personal discussions we were able to get in | ||
contact with people, raise awareness on the issue and also learned a lot ourselves about the landscape | contact with people, raise awareness on the issue and also learned a lot ourselves about the landscape | ||
| − | of opinions.< | + | of opinions. |
| − | + | </p> | |
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
At the same time we gave children and interested adults the chance to take a closer look at our work | At the same time we gave children and interested adults the chance to take a closer look at our work | ||
by | by | ||
| − | letting them experimentally extract DNA from pepper. We used this to also explain to the children what | + | letting them experimentally extract DNA from pepper (figure 1). We used this to also explain to the |
| − | DNA is, what it means to them and what exactly they were doing in each step.< | + | children what |
| − | + | DNA is, what it means to them and what exactly they were doing in each step. | |
| + | </p> | ||
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
Due to the contact and discussion with people from the general society and especially outside of our | Due to the contact and discussion with people from the general society and especially outside of our | ||
university background, the plant breeder festival expanded our knowledge and we therefore see it as a | university background, the plant breeder festival expanded our knowledge and we therefore see it as a | ||
| Line 585: | Line 752: | ||
critics of genetic engineering and took away some lively discussions. In addition, we were able to | critics of genetic engineering and took away some lively discussions. In addition, we were able to | ||
evaluate around 200 questionnaires and further promote our panel discussion. This was demonstrated by | evaluate around 200 questionnaires and further promote our panel discussion. This was demonstrated by | ||
| − | the run on our subsequent panel discussion, where we were happy to recognize some familiar faces from the | + | the run on our subsequent panel discussion, where we were happy to recognize some familiar faces from |
| − | festival.< | + | the |
| − | + | festival. | |
| + | </p> | ||
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
<b>Evaluation</b><br> | <b>Evaluation</b><br> | ||
In our questionnaire, we first had the participants assess their current state of knowledge about | In our questionnaire, we first had the participants assess their current state of knowledge about | ||
| − | genetic engineering and divided the questions into gender, age and basic attitude.< | + | genetic engineering and divided the questions into gender, age and basic attitude. |
| − | + | </p> | |
| − | + | <p style="text-align: justify; margin-bottom: 1em;"> | |
As far as gender is of interest, most of the participants rated themselves in the criteria as | As far as gender is of interest, most of the participants rated themselves in the criteria as | ||
"rather bad" and "medium". Men consider themselves to be better informed than women. Especially in | "rather bad" and "medium". Men consider themselves to be better informed than women. Especially in | ||
| Line 600: | Line 769: | ||
their self-assessment. It is possible that men are more self-confident in this aspect and thus, | their self-assessment. It is possible that men are more self-confident in this aspect and thus, | ||
consider | consider | ||
| − | themselves to be better informed. | + | themselves to be better informed. |
| − | + | ||
</p> | </p> | ||
| − | < | + | <div class="grid" |
| − | < | + | style=" |
| − | <img style=" | + | grid-template-columns: repeat(auto-fit,30vw); |
| − | src=https://static.igem.org/mediawiki/2019/e/e6/T--Marburg--plant-market-sex.jpg | + | "> |
| − | + | ||
| − | + | <figure style="text-align:center;"> | |
| − | < | + | <img style="" |
| − | + | src="https://static.igem.org/mediawiki/2019/e/e6/T--Marburg--plant-market-sex.jpg" | |
| − | + | alt="sex"> | |
| − | + | <figcaption style="max-width: 100%">Fig.2 - Answers analyzed by gender for the question "How | |
| − | + | ||
knowledgable are you about gene | knowledgable are you about gene | ||
editing?".</figcaption> | editing?".</figcaption> | ||
| − | <figcaption | + | </figure> |
| + | <figure style="text-align:center;"> | ||
| + | <img style="" | ||
| + | src="https://static.igem.org/mediawiki/2019/f/f3/T--Marburg--plant-market-age.jpg" | ||
| + | alt="age"> | ||
| + | <figcaption style="max-width: 100%">Fig.3 - Answers analyzed by age for the question "How | ||
knowledgable | knowledgable | ||
are you about gene | are you about gene | ||
editing?".</figcaption> | editing?".</figcaption> | ||
| − | <figcaption | + | </figure> |
| + | <figure style="text-align:center;"> | ||
| + | <img style="" | ||
| + | src="https://static.igem.org/mediawiki/2019/1/1e/T--Marburg--survey_viewvsknowledge.png" | ||
| + | alt="knowledge"> | ||
| + | <figcaption style="max-width: 100%">Fig.4 - Answers analyzed by attitude towards gene editing. | ||
</figcaption> | </figcaption> | ||
| − | </ | + | </figure> |
| − | </ | + | </div> |
<p style="text-align: justify; margin-bottom: 1em;"> | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| − | In the category “age”, the average self-assessment is also "rather bad" and"medium". However, | + | In the category “age”, the average self-assessment is also "rather bad" and "medium". However, |
younger people (18-29) generally considered themselves to be better informed than older people. A | younger people (18-29) generally considered themselves to be better informed than older people. A | ||
| − | problem is that the older generation may have more difficulty accessing information.< | + | problem is that the older generation may have more difficulty accessing information. |
| − | + | </p> | |
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
It is also noticeable that the participants have a positive attitude towards genetic | It is also noticeable that the participants have a positive attitude towards genetic | ||
engineering when they are better informed. This shows that there is not enough information on | engineering when they are better informed. This shows that there is not enough information on | ||
genetic | genetic | ||
| − | engineering and that it does not reach the population.< | + | engineering and that it does not reach the population. |
| − | + | </p> | |
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
The first part of our study reveals two problems: bad accessibility of information to the older | The first part of our study reveals two problems: bad accessibility of information to the older | ||
generation creates a discrepancy between the level of knowledge of younger and older people. As | generation creates a discrepancy between the level of knowledge of younger and older people. As | ||
| Line 644: | Line 823: | ||
as | as | ||
people are obviously better attuned to genetic engineering when they know more about it. In general, | people are obviously better attuned to genetic engineering when they know more about it. In general, | ||
| − | there is a need for better information on the subject and more discussions at a political level.< | + | there is a need for better information on the subject and more discussions at a political level. |
| − | + | </p> | |
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
We then asked whether the participants considered genetic engineering in plants, animals and humans | We then asked whether the participants considered genetic engineering in plants, animals and humans | ||
to be | to be | ||
| − | ethically justifiable.< | + | ethically justifiable. |
| − | + | </p> | |
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
Most people had a positive attitude towards plants. Here, many people probably recognized the | Most people had a positive attitude towards plants. Here, many people probably recognized the | ||
| − | potential, also with regard to the 2050 food problem or resistance to pests.< | + | potential, also with regard to the 2050 food problem or resistance to pests. |
| − | + | </p> | |
| + | <p style="text-align: justify;"> | ||
Surprisingly, many participants are more likely to agree with genetic engineering in | Surprisingly, many participants are more likely to agree with genetic engineering in | ||
humans than in animals. Most people probably think of the potential of genetic engineering to fight | humans than in animals. Most people probably think of the potential of genetic engineering to fight | ||
| Line 667: | Line 849: | ||
<br> | <br> | ||
</p> | </p> | ||
| − | < | + | <div class="grid" |
| − | < | + | style=" |
| − | <img style=" | + | grid-template-columns: repeat(auto-fit,30vw); |
| − | src=https://static.igem.org/mediawiki/2019/9/9b/T--Marburg--gene_editing_plants_pie.png alt="sex"> | + | "> |
| − | < | + | <figure style="text-align:center;"> |
| − | src=https://static.igem.org/mediawiki/2019/b/bf/T--Marburg--gene_editing_animals_pie.png alt="sex"> | + | <img style="" |
| − | < | + | src="https://static.igem.org/mediawiki/2019/9/9b/T--Marburg--gene_editing_plants_pie.png" |
| − | src=https://static.igem.org/mediawiki/2019/9/9b/T--Marburg--gene_editing_plants_pie.png alt="sex"> | + | alt="sex"> |
| − | + | <figcaption style="max-width: 100%;"> | |
| − | + | Fig.5 - Attitude towards gene editing in plants. | |
| − | + | </figcaption> | |
| − | + | </figure> | |
| − | + | <figure style="text-align:center;"> | |
| − | + | <img style="" | |
| − | + | src="https://static.igem.org/mediawiki/2019/b/bf/T--Marburg--gene_editing_animals_pie.png" | |
| − | + | alt="sex"> | |
| − | + | <figcaption style="max-width: 100%;"> | |
| − | + | Fig.6 - Attitude towards gene editing in | |
| − | + | animals. | |
| − | </ | + | </figcaption> |
| − | </ | + | </figure> |
| − | </ | + | <figure style="text-align:center;"> |
| + | <img style="" | ||
| + | src="https://static.igem.org/mediawiki/2019/9/9b/T--Marburg--gene_editing_plants_pie.png" | ||
| + | alt="sex"> | ||
| + | <figcaption style="max-width: 100%;"> | ||
| + | Fig.7 - | ||
| + | Attitude towards gene editing in humans. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </section> | ||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <div class="sub" onclick="popup('panel_discussion')"> | + | <div class="sub" |
| + | onclick="popup('panel_discussion')"> | ||
<div class="sub-header"> | <div class="sub-header"> | ||
<h1> | <h1> | ||
| Line 701: | Line 894: | ||
<div class="sub-content"> | <div class="sub-content"> | ||
<div> | <div> | ||
| − | The panel discussion allowed regional residents to debate with experts from various fields about green genetic engineering. | + | The panel discussion allowed regional residents to debate with experts from various fields about green |
| + | genetic engineering. | ||
</div> | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | <div id="panel_discussion" class="popup"> | + | <div id="panel_discussion" |
| + | class="popup"> | ||
<div class="popup-container"> | <div class="popup-container"> | ||
<div class="popup-header"> | <div class="popup-header"> | ||
<h1 class="title">Genetic engineering panel discussion report: Blessing or curse</h1> | <h1 class="title">Genetic engineering panel discussion report: Blessing or curse</h1> | ||
| − | <button type="button" onclick="hide('panel_discussion')">X</button> | + | <button type="button" |
| + | onclick="hide('panel_discussion')">X</button> | ||
</div> | </div> | ||
| − | <div class="popup-content" style="text-align: justify;"> | + | <div class="popup-content" |
| + | style="text-align: justify;"> | ||
<div> | <div> | ||
<p style="text-align: justify; margin-bottom: 1em;"> | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| Line 741: | Line 938: | ||
</p> | </p> | ||
<figure style="float: left; margin-right: 25px;"> | <figure style="float: left; margin-right: 25px;"> | ||
| − | <img style=" | + | <img style="width: 500px;" |
src="https://static.igem.org/mediawiki/2019/9/9e/T--Marburg--Paneldiscussion_Mikro.jpg" | src="https://static.igem.org/mediawiki/2019/9/9e/T--Marburg--Paneldiscussion_Mikro.jpg" | ||
alt="Placeholder image"> | alt="Placeholder image"> | ||
| − | <figcaption style="max-width: | + | <figcaption style="max-width: 500px;"> |
Fig.1 - Our team member Lars giving the microphone to one of the viewers, that everyone can | Fig.1 - Our team member Lars giving the microphone to one of the viewers, that everyone can | ||
hear his question. | hear his question. | ||
| Line 752: | Line 949: | ||
Most of the panelists agreed that genetic engineering offers opportunities, especially in | Most of the panelists agreed that genetic engineering offers opportunities, especially in | ||
agriculture, to address challenges such as climate change or the nutrition of the growing world | agriculture, to address challenges such as climate change or the nutrition of the growing world | ||
| − | population. According to the introductory statement of Prof. Dr. Andreas Weber from the Cluster of Excellence on Plant | + | population. According to the introductory statement of Prof. Dr. Andreas Weber from the Cluster of |
| + | Excellence on Plant | ||
Science at Heinrich-Heine University in Düsseldorf, methods of genetic | Science at Heinrich-Heine University in Düsseldorf, methods of genetic | ||
engineering such as CRISPR are already established tools and have no cause for concern, particularly | engineering such as CRISPR are already established tools and have no cause for concern, particularly | ||
| − | in fundamental research. But subsequently, modified organisms cannot be tested in the field under natural | + | in fundamental research. But subsequently, modified organisms cannot be tested in the field under |
| + | natural | ||
conditions. However, the objective of testing and checking modified plants is a problem, because | conditions. However, the objective of testing and checking modified plants is a problem, because | ||
often independent testing organisations do not have access to certified seeds and the complete | often independent testing organisations do not have access to certified seeds and the complete | ||
| Line 766: | Line 965: | ||
different background, here represented by ethics professor Dr. Friedemann Voigt, put the consumers in | different background, here represented by ethics professor Dr. Friedemann Voigt, put the consumers in | ||
the foreground and demanded their right to know how a food is produced. This led the discussion | the foreground and demanded their right to know how a food is produced. This led the discussion | ||
| − | further in the direction of the effects of genetically modified animal feed on farm animals and the human | + | further in the direction of the effects of genetically modified animal feed on farm animals and the |
| + | human | ||
consumer. At this point, Dr. Christoph Then criticised the sharp decline in risk research, which | consumer. At this point, Dr. Christoph Then criticised the sharp decline in risk research, which | ||
pursues society's protective goals with regard to health.<br> | pursues society's protective goals with regard to health.<br> | ||
| Line 777: | Line 977: | ||
approval of the audience, which was expressed by strong applause. Transgenic plants could not | approval of the audience, which was expressed by strong applause. Transgenic plants could not | ||
develop in a natural way and should therefore not be used in agriculture. At the moment, however, | develop in a natural way and should therefore not be used in agriculture. At the moment, however, | ||
| − | the use of genetically modified plants in organic farming is also not in compliance with | + | the use of genetically modified plants in organic farming is also not in compliance with |
regulations, according to Freya Schäfer of FiBL (Research Institute of Organic Agriculture).<br> | regulations, according to Freya Schäfer of FiBL (Research Institute of Organic Agriculture).<br> | ||
<br> | <br> | ||
</p> | </p> | ||
<figure style="float: right; margin-left: 25px;"> | <figure style="float: right; margin-left: 25px;"> | ||
| − | <img style=" | + | <img style="width: 500px;" |
src="https://static.igem.org/mediawiki/2019/e/e1/T--Marburg--Paneldiscussion_Podium.jpg" | src="https://static.igem.org/mediawiki/2019/e/e1/T--Marburg--Paneldiscussion_Podium.jpg" | ||
alt="Placeholder image"> | alt="Placeholder image"> | ||
| − | <figcaption> | + | <figcaption style="max-width: 500px;"> |
| − | Fig.2 - Recording of the panelists in the middle of arguing with the viewers. Members of the panel are (left to right):<br> Paula Müller, Prof. Dr. Friedemann Voigt, Dr. Claus Kremoser, Daniel Stukenberg, Freya Schäfer, Dr. Christoph Then,<br> Prof. Dr. Andreas Weber and Michael Lange. | + | Fig.2 - Recording of the panelists in the middle of arguing with the viewers. Members of the panel |
| + | are (left to right):<br> Paula Müller, Prof. Dr. Friedemann Voigt, Dr. Claus Kremoser, Daniel | ||
| + | Stukenberg, Freya Schäfer, Dr. Christoph Then,<br> Prof. Dr. Andreas Weber and Michael Lange. | ||
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| Line 813: | Line 1,015: | ||
</div> | </div> | ||
</section> | </section> | ||
| − | + | <hr> | |
| − | + | <section class="section"> | |
| − | + | <h1 class="title">Integrated Human Practices</h1> | |
| − | + | <div class="grid"> | |
| − | + | <div class="sub" | |
| − | + | onclick="popup('cyano_biotech')"> | |
| − | + | <div class="sub-header"> | |
| − | + | <h1> | |
| − | + | C Y A N O<br> | |
| − | + | B I O T E C H | |
| − | + | </h1> | |
| − | + | <hr> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
| − | + | <div class="sub-content"> | |
| − | + | <div> | |
| − | + | Influencing our project on many levels: growth curves, terminators and well plate cultivation. | |
| − | + | </div> | |
| − | <div class=" | + | |
| − | < | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div id="cyano_biotech" | |
| − | + | class="popup"> | |
| − | <div | + | <div class="popup-container"> |
| − | + | <div class="popup-header"> | |
| − | + | <h1 class="title">Cyano Biotech</h1> | |
| − | + | <button type="button" | |
| − | + | onclick="hide('cyano_biotech')">X</button> | |
| − | + | </div> | |
| − | + | <div class="popup-content" | |
| − | + | style="text-align: justify;"> | |
| − | + | <section class="section"> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<p style="text-align: justify; margin-bottom: 1em;"> | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| − | + | Cyano Biotech is one of the leading, top edge companies to work with sustainable and | |
| − | + | product-oriented | |
| − | + | phototrophic bacteria in Germany. As a shining figure in cyanobacterial metabolic engineering, the | |
| − | + | CEO | |
| − | + | of Cyano Biotech Dr. Dan Enke talked with us about the possibilities of our project. His feedback | |
| + | led | ||
| + | us to the design of adjusting specific parameters in our <a | ||
| + | href="https://2019.igem.org/Team:Marburg/Model#growth_curve_model" | ||
| + | target="_blank">growth experiments</a> and to the integration of | ||
| + | our | ||
| + | <a href="https://2019.igem.org/Team:Marburg/Model#terminator_model" | ||
| + | target="_blank">terminator library</a>. Through our talk with a potential end user of our Green | ||
| + | Expansion | ||
| + | and our | ||
| + | engineered | ||
| + | strains, we achieved a more “real-world” focus of our project.<br> | ||
| + | <br> | ||
| + | As our project was in its beginning, one thing was clear: high throughput methods are essential for | ||
| + | any | ||
| + | viable chassis in Synthetic Biology. Sadly, we quickly noticed obstacles in our way, such as | ||
| + | inhibited | ||
| + | growth in our plates compared to flasks. In consequence of that, we looked for help and who would be | ||
| + | better, than an expert on that specific field? We talked to Dan Enke and he kindly provided us with | ||
| + | data in his own well plate experiments. Soon we noticed, that he inoculated cultures several times | ||
| + | in a | ||
| + | row out of the exponential phase, leading to a huge boost in doubling times. Through this support we | ||
| + | were able to design and conduct a growth curve with as much as five precultures.<br> | ||
<br> | <br> | ||
| + | Aside from that, we talked about another facet of our project, cyanobacterial terminators. Being | ||
| + | overread in | ||
| + | cyano´s, both metaphorically and for real, they are one of the most important | ||
| + | regulatory genetic elements. Yet another expert in the | ||
| + | field of cyanobacteria was pleased of our idea to set on the search for the best terminator and | ||
| + | strongly encouraged us to utilize the Green Expansion to its full extent. <br> | ||
</p> | </p> | ||
| − | <figure style=" | + | <figure style="text-align: center"> |
| − | <img style="height: | + | <img style="height: 60ex; width: 50ex" |
| − | src="https://static.igem.org/mediawiki/2019/ | + | src="https://static.igem.org/mediawiki/2019/2/2c/T--Marburg--CyanoBiotech.png" |
| − | <figcaption style=" | + | alt="CyanoBiotech Skype Call with Dr. Dan Enke"> |
| − | Fig.1 - | + | <figcaption style="text-align: center"> |
| − | + | Fig.1 - Skype Call with Dr. Dan Enke from CyanoBiotech. | |
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | </ | + | |
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div class="sub" | |
| − | + | onclick="popup('prof_wilde')"> | |
| − | + | <div class="sub-header"> | |
| − | + | <h1> | |
| − | + | P R O F.   D R.<br> | |
| − | + | A N N E G R E T   W I L D E | |
| − | + | </h1> | |
| + | <hr> | ||
| + | </div> | ||
| + | <div class="sub-content"> | ||
| + | <div> | ||
| + | Cultivation expertise from leading cyano scientist Prof. Wilde. | ||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
| − | <div class=" | + | <div id="prof_wilde" |
| − | <div> | + | class="popup"> |
| − | + | <div class="popup-container"> | |
| + | <div class="popup-header"> | ||
| + | <h1 class="title">Cultivation expertise from leading cyano scientist Prof. Wilde</h1> | ||
| + | <button type="button" | ||
| + | onclick="hide('prof_wilde')">X</button> | ||
| + | </div> | ||
| + | <div class="popup-content" | ||
| + | style="text-align: justify;"> | ||
| + | <section class="section"> | ||
| + | <div> | ||
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| + | While working with the cyanobacterium <i>Synechococcus elongatus</i> UTEX 2973 we noticed very | ||
| + | quickly a | ||
| + | lack of | ||
| + | standardization in the field of Synthetic Biology. To tackle this huge problem, we | ||
| + | decided to focus, as one of our main goals, on standardization to make scientific results more | ||
| + | comparable. Therefore, we worked on standardizing light measurement, cultivating parameters | ||
| + | (temperature, CO2, rpm, …) and the cultivation media for cyanobacteria, especially UTEX 2973.<br> | ||
| + | <br> | ||
| + | </p> | ||
| + | <figure style="float: left; margin-right: 25px;"> | ||
| + | <img style="height: 500px; width: 650px" | ||
| + | src="https://static.igem.org/mediawiki/2019/b/bb/T--Marburg--Wilde_and_us.jpg" | ||
| + | alt="Placeholder image"> | ||
| + | <figcaption style="max-width: 650px"> | ||
| + | Fig.1 - Prof. Dr. Annegret Wilde and our team members Vinca, Marian and Robin in | ||
| + | the botanical garden of Freiburg. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| + | During the theoretical planning of our project we contacted scientists in research and industry | ||
| + | that | ||
| + | are specialists for cyanobacteria. As a result, Prof. Dr. Annegret Wilde (Institute of Biology, | ||
| + | Albert-Ludwigs-University Freiburg) answered our call and was very interested in advising us in | ||
| + | regards to our projects. In order to give us an introduction to the handling of cyanobacteria, she | ||
| + | invited us to her institute at the University of Freiburg on 5th and 6th June 2019. In a short and | ||
| + | focused internship we were able to quickly gain a set of core competencies regarding sterile | ||
| + | inoculation, streaking of cyano cultures and further information regarding the cultivation | ||
| + | conditions | ||
| + | which we applied to our strain.<br> | ||
| + | <br> | ||
| + | In addition, we also learned that the measurement of light intensity is an important topic. There | ||
| + | is | ||
| + | a | ||
| + | variety of measuring instruments and different methods for each, which means that information on | ||
| + | light | ||
| + | intensities should be viewed with caution. For cyanobacteria such as <i>Synechococcus | ||
| + | elongatus</i> UTEX | ||
| + | 2973 | ||
| + | light intensity plays a decisive role, which is why we analysed differences between a variety of | ||
| + | instruments and methods to establish a standard for <a | ||
| + | href="https://2019.igem.org/Team:Marburg/Measurement#light_measurement" | ||
| + | target="_blank">light measurement</a> based on our results .<br> | ||
| + | <br> | ||
| + | In further discussions about our Marburg Collection 2.0 we were recommended to take a very close | ||
| + | look | ||
| + | at <a href="https://2019.igem.org/Team:Marburg/Model#terminator_model" | ||
| + | target="_blank">terminators</a> as they oddly enough have an effect on the transcription of | ||
| + | upstream | ||
| + | genes. As a | ||
| + | result, we decided to take a closer look at that and investigated their effects. We thank | ||
| + | Professor | ||
| + | A. | ||
| + | Wilde for her input, her invitation to Freiburg and her recommendations that guided us in our | ||
| + | project. | ||
| + | <b></b><br> | ||
| + | <br> | ||
| + | </p> | ||
| + | </section> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div class="sub" | |
| − | + | onclick="popup('doulix')"> | |
| − | + | <div class="sub-header"> | |
| − | <div class=" | + | <h1> |
| − | <h1 | + | D O U L I X<br> |
| − | < | + | </h1> |
| + | <hr> | ||
</div> | </div> | ||
| − | <div class="popup-content" style="text-align: justify;"> | + | <div class="sub-content"> |
| − | + | <div> | |
| − | + | Confirmation for real case use for our colony picking project. | |
| − | + | </div> | |
| − | + | </div> | |
| − | + | </div> | |
| − | + | <div id="doulix" | |
| − | + | class="popup"> | |
| − | + | <div class="popup-container"> | |
| − | + | <div class="popup-header"> | |
| − | + | <h1 class="title">Doulix</h1> | |
| − | + | <button type="button" | |
| − | + | onclick="hide('doulix')">X</button> | |
| − | + | </div> | |
| − | + | <div class="popup-content" | |
| − | + | style="text-align: justify;"> | |
| − | + | <section class="section"> | |
| − | + | <div> | |
| − | + | <p style="text-align: center; margin-bottom: 1em;"> | |
| − | + | “My major bottleneck is colony picking.” - <b>Davide De Lucrezia</b>, Managing Director of | |
| − | + | Doulix</p> | |
<br> | <br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<br> | <br> | ||
| − | That is where our team came into play. We decided to take this advice to our heart and started to | + | <p style="text-align: justify; margin-bottom: 1em;"> |
| − | + | The vision of automating colony picking has existed now for many years. Big companies like Tecan, | |
| − | + | Singer Instruments or Hudson Robotics invented robots those are able to identify and pick | |
| − | + | colonies. | |
| − | + | The problem is that they cost a fortune, starting at 50,000€ up to 100,000€ or even more. Although | |
| − | + | it is highly desired, a low cost solution is still missing for this tedious task. Start-ups like | |
| − | + | Doulix are currently automating such workflows in their laboratories, but they do not have the | |
| − | + | funds | |
| − | + | to finance a state-of-the-art colony picking robot. By now every single step has to be performed | |
| − | + | manually, draining resources from other departments, which actually should be paid more attention | |
| − | + | to. “Colony picking is a bottleneck in every of part our workflows” says Davide De Lucrezia, the | |
| − | + | founder and managing director of Doulix. Doulix focuses on developing innovative technologies for | |
| − | + | scientists to simplify their work, especially in the field of Synthetic Biology, and they are | |
| − | + | currently planning on establishing Opentrons OT-2 in their lab to automate the most part of their | |
| − | + | workflows.<br> | |
| − | + | <br> | |
| − | + | During an online conference with Davide De Lucrezia, Sota Hirano, and Alessandro Filisetti from | |
| − | + | Doulix, Davide De Lucrezia suggested that turning the OT-2 into a colony picker as a project would | |
| − | + | be | |
| − | + | really | |
| − | + | interesting. To have a fully trained, ready to use package to turn the OT-2 into a colony picker | |
| − | + | would enhance the workflow at Doulix tremendously. Nevertheless, to suit the user's needs as well | |
| − | + | as | |
| − | + | to get this job done in the spirit of Opentrons, installing the needed add-ons should be as | |
| − | + | modular | |
| − | + | and flexible as possible and designed so that “even a biologist” without technical knowledge or | |
| − | + | programming skills would be able to install and use them.<br> | |
| − | + | <br> | |
| − | + | That is where our team came into play. We decided to take this advice to our heart and started to | |
| − | + | work out what was needed to turn the OT-2 into a fully automated colony picking robot.<br> | |
| − | + | <br> | |
| − | + | </p> | |
| − | + | <figure style="float: right; margin-left: 25px;"> | |
| − | + | <img style="height: 400px; width: 600px" | |
| − | + | src="https://static.igem.org/mediawiki/2019/f/f8/T--Marburg--doulix.png" | |
| − | + | alt="Connections between Opentrons, Promega and QInstruments"> | |
| − | + | <figcaption style="max-width: 600px"> | |
| − | + | Fig.1 - iGEM team Marburg 2019 meeting with Davide De Lucrezia (left), Sota Hirano (middle) and | |
| − | + | Alessandro Filisetti (right) in a Zoom teleconference to discuss advanced automated workflows in | |
| − | + | the lab. | |
| − | + | </figcaption> | |
| − | + | </figure> | |
| − | + | <p style="text-align: justify; margin-bottom: 1em;"> | |
| − | + | One of the first big design questions was whether we wanted to hardcode an image recognition | |
| − | + | software for the colony detection or if it was a better choice to train a data hungry but - given | |
| − | + | proper and enough training data - more accurate and scalable artificificial intelligence based | |
| − | + | colony detection. Kristin Ellis, the director of strategic initiatives at <a | |
| − | + | href="https://2019.igem.org/Team:Marburg/Human_Practices#opentrons_keoni_gandall" | |
| − | + | target="_blank">Opentrons referred us to | |
| − | + | Keoni Gandall</a> from Stanford, a well known tinker of the OT-2 for more unconventional | |
| + | applications. | ||
| + | He is building a colony picking system himself, however he chose not to rely on an AI. He | ||
| + | recommended us to go with AI as he thinks his approach is very prone to changes in parameters. If | ||
| + | many | ||
| + | different users want to utilize the same system, a flexible software is required that can take | ||
| + | environmental changes into account. We decided to opt for maximum flexibility by <a | ||
| + | href="https://2019.igem.org/Team:Marburg/Colony_Picking" | ||
| + | target="_blank">working with an | ||
| + | AI</a>.<br> | ||
| + | <br> | ||
| + | Now that we had an idea of the required software we started to design modular<a | ||
| + | href="https://2019.igem.org/Team:Marburg/Hardware" | ||
| + | target="_blank"> hardware</a> to overcome | ||
| + | potential problems in a fully automated workflow in the OT-2. To illuminate the agar plates in the | ||
| + | right | ||
| + | way without any distortions we | ||
| + | engineered a light table that distributes light equally over the plate.<br> | ||
| + | <br> | ||
| + | To give an “eyesight” to the OT-2 we mounted a Raspberry Pi 4 and an ArduCAM on the OT-2 arm. For | ||
| + | a | ||
| + | better accessibility we created our Graphical User Interface for Directed Engineering <a | ||
| + | href="https://2019.igem.org/Team:Marburg/Colony_Picking" | ||
| + | target="_blank">(GUIDE)</a>. We | ||
| + | designed our GUI in a way that will enable every user to train their own AI with their own | ||
| + | training | ||
| + | data set so that the AI can be optimized for each specific situation. Moreover the GUI will also | ||
| + | enable access to users that are not trained in computer/ software engineering.<br> | ||
| + | <br> | ||
| + | Now that we gave our robot the ability to see, to think and to communicate with us, nothing stood | ||
| + | in | ||
| + | the way of our own colony picker. We are now able to turn the OT-2 into a colony picker costing | ||
| + | <a href="https://2019.igem.org/Team:Marburg/Colony_Picking" | ||
| + | target="_blank">below $300</a>. Moreover, for companies, teams or | ||
| + | groups who do not own an OT-2 yet, we were able to reduce the costs for purchasing a colony picker | ||
| + | by 90-95%, compared to the listed market prices for traditional colony pickers.<br> | ||
| + | <br> | ||
| + | Finally we contacted Doulix the second time to discuss in more details about our project. They | ||
| + | approved it and gave further suggestions such as “live training” so that the AI will continue to | ||
| + | learn as it is being used to pick up colonies. Not only will this improve the AI gradually but it | ||
| + | will also adapt to the specific needs of each user. This leaves a lot of room to improve the | ||
| + | project | ||
| + | in the future.<br> | ||
| + | <br> | ||
| + | </p> | ||
| + | </section> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div class="sub" | |
| − | + | onclick="popup('standardization')"> | |
| − | + | <div class="sub-header"> | |
| − | + | <h1 style="font-size: .9rem !important;"> | |
| − | + | S T A N D A R D I Z A T I O N | |
| − | + | </h1> | |
| − | + | <hr> | |
| + | </div> | ||
| + | <div class="sub-content"> | ||
| + | <div> | ||
| + | Realizing that we are not the only ones struggeling with no cyano research standarization... | ||
| + | </div> | ||
| + | </div> | ||
</div> | </div> | ||
| − | <div class=" | + | <div id="standardization" |
| − | <div> | + | class="popup"> |
| − | + | <div class="popup-container"> | |
| + | <div class="popup-header"> | ||
| + | <h1 class="title">Standardization in Cyanocommunity</h1> | ||
| + | <button type="button" | ||
| + | onclick="hide('standardization')">X</button> | ||
| + | </div> | ||
| + | <div class="popup-content" | ||
| + | style="text-align: justify;"> | ||
| + | <section class="section"> | ||
| + | <div> | ||
| + | <figure style="float: right; margin-left: 25px;"> | ||
| + | <img style="width: 500px;" | ||
| + | src="https://static.igem.org/mediawiki/2019/5/5c/T--Marburg--CyanoCommunity_BG11.png" | ||
| + | alt="Placeholder image"> | ||
| + | <figcaption style="max-width: 500px;"> | ||
| + | Fig.1 - Growth Curve UTEX 2973 in BG11 and BGM. Medium A: BGM, Medium B: BG11 from Uni Marburg, | ||
| + | Medium C: BG11 from Uni Tübingen, Medium D: BG 11 from Uni Düsseldorf. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| + | During our visit at the Cyano Conference 2019 in Tübingen we recognized a need for standardization | ||
| + | in | ||
| + | this community. We asked people to send us their <a | ||
| + | href="https://2019.igem.org/Team:Marburg/Experiments#protocols" | ||
| + | tagret="_blank">BG-11 recipes</a> and | ||
| + | surprisingly | ||
| + | received several different versions. The community at the conference was aware of the missing | ||
| + | standardization and we received very positive feedback for our efforts. Fixed standards are | ||
| + | essential | ||
| + | for reproducibility of results, especially the preparation of media and buffers but no one is | ||
| + | investing time to set a standard. After the cyano conference we stayed in contact with <a | ||
| + | href="https://2019.igem.org/Team:Marburg/Human_Practices#nicolas_schmelling" | ||
| + | target="_blank"> Nicolas | ||
| + | Schmelling</a>, coordinator of the bachelor program at <a href="" | ||
| + | target="_blank">CEPLAS</a>. During his PhD he was working on | ||
| + | establishing more standards in the cyano community. He tried to establish protocols and collected | ||
| + | different methods and recipes to establish a standard for all.<br> | ||
| + | <br> | ||
| + | After he clarified to us the importance of comparable media in context of standards, we started to | ||
| + | collect different BG11 recipes and compared them in growing experiments. Figure 1 | ||
| + | depicts our results and shows the impact of different recipes for media.<br> | ||
| + | In our experiment we could show that there is a significant difference between the different BG11 | ||
| + | recipes despite their relative similarity. During our complete project we were working on the | ||
| + | standardization for <a href="https://2019.igem.org/Team:Marburg/Measurement" | ||
| + | target="_blank"> light intensity, media and different cultivating parameters</a>. We made it our | ||
| + | destiny | ||
| + | to make the first step into the beginning of | ||
| + | standardization in | ||
| + | the cyano community by providing <i>Synechococcus elongatus</i> with standardized | ||
| + | parameters. We | ||
| + | were able to find the optimal growing conditions for UTEX 2973 and could show that creating a | ||
| + | standard | ||
| + | in measurement and methods is really important to have comparable results. With our project we | ||
| + | hope | ||
| + | that we could set a first step into standardization, so that the future cyano community will have | ||
| + | standardized and comparable results.<br> | ||
| + | </p> | ||
| + | </div> | ||
| + | </section> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div class="sub" | |
| − | + | onclick="popup('cyano_conference')"> | |
| − | + | <div class="sub-header"> | |
| − | <div class=" | + | <h1 style="font-size: .9rem !important;"> |
| − | <h1 | + | C Y A N O <br> |
| − | < | + | C O N F E R E N C E   2 0 1 9 |
| + | </h1> | ||
| + | <hr> | ||
</div> | </div> | ||
| − | <div class=" | + | <div class="sub-content"> |
| − | < | + | <div> |
| − | + | Knowledge exchange with established cyanobacteria researchers. | |
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div id="cyano_conference" | |
| − | + | class="popup"> | |
| − | + | <div class="popup-container"> | |
| − | + | <div class="popup-header"> | |
| − | + | <h1 class="title">The CYANO Conference 2019 in Tübingen</h1> | |
| − | + | <button type="button" | |
| − | + | onclick="hide('cyano_conference')">X</button> | |
| − | + | </div> | |
| − | + | <div class="popup-content" | |
| − | + | style="text-align: justify;"> | |
| − | + | <section class="section"> | |
| − | + | <div> | |
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| + | From September 11th to September 13th we attended the CYANO Conference 2019 in Tuebingen funded by | ||
| + | the <a href="https://vaam.de" | ||
| + | target="_blank">VAAM</a> (Vereinigung für Allgemeine und Angewandte Mikrobiologie). During the | ||
| + | poster | ||
| + | sessions we | ||
| + | took the chance to present our project and how we revolutionize the upcoming work on phototrophic | ||
| + | organisms. Therefore, we gained great feedback from the participants, which showed huge interest | ||
| + | in | ||
| + | <a href="https://2019.igem.org/Team:Marburg/Results#marburg_collection" | ||
| + | target="_blank">our toolbox specified for cyanobacteria</a>. Our Synthetic Biology approaches | ||
| + | encountered the thinking | ||
| + | of classical cyanobacterial research which lead to interesting discussions from which we gained a | ||
| + | lot of input. Furthermore, the leading experts of cyanobacteria offered talks where we learned how | ||
| + | to modify working on <i>Synechococcus elongatus</i>.</p> | ||
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| + | We were especially interested in the discussions about methods. We soon realized that the | ||
| + | cyanobacterial scene has no standardized protocols for daily laboratory practices and they are | ||
| + | also | ||
| + | aware of that issue. This started with debates about the media composition of BG-11 media but also | ||
| + | concerned issues like standardized evaluation of light conditions. With our project for | ||
| + | standardizing growth conditions and providing a part collection we tackle these major issues for | ||
| + | scientists studying phototrophic organisms.</p> | ||
| + | |||
| + | <figure style="float: left; margin-right: 25px;"> | ||
| + | <img style="height: 600px; width: 1500px" | ||
| + | src="https://static.igem.org/mediawiki/2019/f/fa/T--Marburg--CyanoConference_Grouptphoto.png" | ||
| + | alt="Placeholder image"> | ||
| + | <figcaption style="max-width: 1500px"> | ||
| + | Fig.1 - Group photo with our team members Vinca, Hinrik, Jana and all experts of the Cyano | ||
| + | Conference. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </section> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div class="sub" | |
| − | + | onclick="popup('james_golden')"> | |
| − | + | <div class="sub-header"> | |
| − | <div class=" | + | <h1 style="font-size: .8rem !important;"> |
| − | <h1 | + | E X P E R T   O N   C Y A N O S :<br> |
| − | < | + | J A M E S   G O L D E N |
| + | </h1> | ||
| + | <hr> | ||
</div> | </div> | ||
| − | <div class=" | + | <div class="sub-content"> |
| − | < | + | <div> |
| − | + | Defining the absent standardization to discrepancies in light, optical denisty measurement and more. | |
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div id="james_golden" | |
| − | + | class="popup"> | |
| − | + | <div class="popup-container"> | |
| − | + | <div class="popup-header"> | |
| − | + | <h1 class="title">Expert on Cyanos - James Golden</h1> | |
| − | + | <button type="button" | |
| − | + | onclick="hide('james_golden')">X</button> | |
| − | + | </div> | |
| − | + | <div class="popup-content" | |
| − | + | style="text-align: justify;"> | |
| − | + | <section class="section"> | |
| − | + | <div> | |
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| + | While diving deeper and deeper into the ocean of possibilities that cyanobacteria have to offer we | ||
| + | noticed a few inconsistencies in literature.<br> | ||
| + | <br> | ||
| + | BG11 media is commonly used in cyanobacterial research, but the exact composition seemed to be | ||
| + | different across every second paper we read. Optical densities are more frequently measured at a | ||
| + | wavelength of 730nm, though 750nm seems to be the better choice. For <i>Synechococcus | ||
| + | elongatus</i> | ||
| + | UTEX 2973 | ||
| + | the “optimal growth conditions” according to literature are often quite different; some state 38°C | ||
| + | and | ||
| + | 500µE at a CO2 level of 3% fits best, others prefer 41°C and 1500µE with 5% CO2 concentration. | ||
| + | <a href="https://2019.igem.org/Measurement#light_measurement" | ||
| + | target="_blank">But how are these light intensities measured?</a> With a planar device or a | ||
| + | spherical | ||
| + | one? We have not | ||
| + | seen this being explained in literature.<br> | ||
| + | <br> | ||
| + | As all of these things have an incredibly huge impact on various different experiments we saw the | ||
| + | need | ||
| + | to find a standardized answer to our questions, reaching out to as many experts in this field as | ||
| + | we | ||
| + | could reach - whether it was industry or research. | ||
| + | One of the leading laboratories working with cyanobacteria is the Golden Lab of the UC San Diego. | ||
| + | Susan Golden and her husband James W. Golden have both been working with cyanobacteria for quite | ||
| + | some | ||
| + | time, now with a stronger focus on their use for industrial purposes. | ||
| + | We set up a Skype call with them, but sadly Susan Golden was not able to join us on short | ||
| + | notice.<br> | ||
| + | <br> | ||
| + | During our talk with James W. Golden we laid open our concerns about the cyanobacterial community | ||
| + | and | ||
| + | he quickly supported our train of thoughts, as he himself noticed a lack of standardization. He | ||
| + | assured us that this is a hot topic in this field of research, as many do not seem to care enough | ||
| + | about the reproducibility of their data and encouraged us to continue with our efforts.<br> | ||
| + | <br> | ||
| + | More accurately, he talked with us about why he thinks there is still no clear decision on whether | ||
| + | to | ||
| + | measure optical densities at 730nm or 750nm: It might be a technical problem, as many photometers | ||
| + | are | ||
| + | simply not able to measure wavelengths of 750nm. In contrast, he mentioned that 750nm would be the | ||
| + | more optimal way, as it proves to minimize absorbance from pigments in cyanobacterial cells, | ||
| + | presenting more accurate data. This confronted us with a conflicting decision: Would it be better | ||
| + | to | ||
| + | use the more accurate 750nm or 730nm, as the latter would allow more labs all over the world to | ||
| + | measure in the same way.<br> | ||
| + | <br> | ||
| + | This was one of the key factors that led us to measure the whole spectrum of our cultures for our | ||
| + | growth curves, as this would provide a larger dataset, awarding us not just with 730nm and 750nm | ||
| + | data, | ||
| + | but also the possibility to check if the spectrum shows normal behavior, from which one could | ||
| + | conclude | ||
| + | how healthy the cultures are. | ||
| + | </p> | ||
| + | <figure style="float: right; margin-left: 25px;"> | ||
| + | <img style="width: 500px;" | ||
| + | src="https://static.igem.org/mediawiki/2019/e/e8/T--Marburg--JG_dif_measurements.png" | ||
| + | alt="JG dif media"> | ||
| + | <figcaption style="max-width: 500px;"> | ||
| + | Fig.1 - Growth of <i>S. elongatus</i> UTEX 2973 at 1500µE measured with different methods. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| + | In the beginning we measured the light intensity of our incubators with a planar | ||
| + | measurement device - the only one available for us. Talking to James Golden we realized that we | ||
| + | should | ||
| + | try to get hold of a spherical measurement device, as he assured us that this is the way to | ||
| + | generate | ||
| + | more accurate data, leading to a more reproducible setup - exactly what we were aiming for.<br> | ||
| + | <br> | ||
| + | After receiving such a device from Biospherical Instruments, we again implemented the feedback we | ||
| + | got and | ||
| + | measured growth curves. | ||
| + | One with cultures at 1500µE measured with a spherical device and one with 1500µE measured with a | ||
| + | planar device, where the measured intensities were converted to theoretically spherical values | ||
| + | with | ||
| + | a | ||
| + | conversion chart offered to us by Prof. Dr. Annegret Wilde from the University of Freiburg.<br> | ||
| + | <br> | ||
| + | These experiments were a huge step in our project, as they heavily influenced the way we cultured | ||
| + | our | ||
| + | cyanobacteria, not only drastically improving their growth, but also clearly demonstrating how | ||
| + | flawed | ||
| + | certain measurements can be. | ||
| + | We would never have been able to reach the fast doubling times we achieve now without this crucial | ||
| + | input and as this will be the case for others too, we made it our mission to keep on stressing the | ||
| + | importance of this way of measurement whenever we reach out to the scientific community. | ||
| + | Again, thank you very much Prof. Dr. James W. Golden for your invaluable contribution! | ||
| + | </p> | ||
| + | </div> | ||
| + | </section> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div class="sub" | |
| − | + | onclick="popup('nicolas_schmelling')"> | |
| − | + | <div class="sub-header"> | |
| − | <div class=" | + | <h1 style="font-size: .8rem !important;"> |
| − | <h1 | + | E X P E R T   O N   C Y A N O S:<br> |
| − | < | + | N I C O L A S  S C H M E L L I N G |
| + | </h1> | ||
| + | <hr> | ||
</div> | </div> | ||
| − | <div class=" | + | <div class="sub-content"> |
| − | < | + | <div> |
| − | + | And soon, we could add growth media to the growing list of not existing standardization. | |
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div id="nicolas_schmelling" | |
| − | + | class="popup"> | |
| − | + | <div class="popup-container"> | |
| − | + | <div class="popup-header"> | |
| − | + | <h1 class="title">Expert on Cyanos - Nicolas Schmelling</h1> | |
| − | + | <button type="button" | |
| − | + | onclick="hide('nicolas_schmelling')">X</button> | |
| − | + | </div> | |
| − | + | <div class="popup-content" | |
| − | + | style="text-align: justify;"> | |
| − | + | <section class="section"> | |
| − | + | <div> | |
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| + | The composition of BG11 media is another important issue we were able to discuss with Nicolas | ||
| + | Schmelling. | ||
| + | While working in our own lab we already got the notion that not all BG11 media are prepared in the | ||
| + | same | ||
| + | way, which is the reason why we kindly asked other researchers from the cyano community- like | ||
| + | James Golden | ||
| + | - to send us their recipes. In order to compare the various ways the BG11 media can be prepared, | ||
| + | we tried | ||
| + | those recipes and measured growth curves to find the perfect fit.<br> | ||
| + | </p> | ||
| + | <figure style="text-align: center"> | ||
| + | <img style="height: 50ex; width: 75ex" | ||
| + | src="https://static.igem.org/mediawiki/2019/8/85/T--Marburg--JG_dif_media.png" | ||
| + | alt="JG dif media"> | ||
| + | <img style="height: 50ex; width: 75ex" | ||
| + | src="https://static.igem.org/mediawiki/2019/9/94/T--Marburg--JG_dif_media_log.png" | ||
| + | alt="JG dif media log"> | ||
| + | <figcaption style="max-width: 150ex;"> | ||
| + | Fig.1 - (Left) Growth of S. elongatus UTEX 2973 in different media. (Right) Growth of S. | ||
| + | elongatus | ||
| + | UTEX 2973 in different media (log-scale). | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| + | It was clear that the growth of our cultures was comparably fast at the beginning no matter what | ||
| + | media | ||
| + | was used, but one of them stood out: BGM - it enabled faster growth at higher ODs, allowing | ||
| + | cultures | ||
| + | to reach double the OD of other cultures after the same time.<br> | ||
| + | <br> | ||
| + | We are certain that having the same ideal medium throughout different cyano labs is not just | ||
| + | elemental | ||
| + | for optimal growth, but also vital for comparability, as trying to reproduce the growth conditions | ||
| + | of | ||
| + | papers can be quite tricky when it is not clear what exact medium was used and how it was | ||
| + | prepared.<br> | ||
| + | <br> | ||
| + | </p> | ||
| + | <figure style="text-align: center"> | ||
| + | <img style="height: 60ex; width: 100ex" | ||
| + | src="https://static.igem.org/mediawiki/2019/0/07/T--Marburg--NicolasSchmelling.png" | ||
| + | alt="Extracting pepper"> | ||
| + | <figcaption style="text-align: center"> | ||
| + | Fig.1 - Skype call with Nicolas Schmelling. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | </div> | ||
| + | </section> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div class="sub" | |
| − | + | onclick="popup('opentrons_keoni_gandall')"> | |
| − | + | <div class="sub-header"> | |
| − | <div class=" | + | <h1 style="font-size: .9rem !important;"> |
| − | <h1 | + | O P E N T R O N S<br> |
| − | < | + | &   K E O N I   G A N D A L L |
| + | </h1> | ||
| + | <hr> | ||
</div> | </div> | ||
| − | <div class=" | + | <div class="sub-content"> |
| − | < | + | <div> |
| − | + | Initial inspirations for our colony picking project. | |
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div id="opentrons_keoni_gandall" | |
| − | + | class="popup"> | |
| − | + | <div class="popup-container"> | |
| − | + | <div class="popup-header"> | |
| − | + | <h1 class="title">Opentrons + Keoni Gandall</h1> | |
| − | + | <button type="button" | |
| − | + | onclick="hide('opentrons_keoni_gandall')">X</button> | |
| − | + | </div> | |
| − | + | <div class="popup-content" | |
| − | + | style="text-align: justify;"> | |
| − | + | <section class="section"> | |
| − | + | <div> | |
| + | <p style="text-align: center; margin-bottom: 1em;"> | ||
| + | “A <a href="https://2019.igem.org/Team:Marburg/Colony_Picking" | ||
| + | target="_blank">colony picking module</a> for the OT-2 will be a great help” - <b>Keoni Gandall | ||
| + | </b></p> | ||
| + | <p style="text-align: justify; margin-bottom: 1em;"> | ||
| + | We started with the colony picking project back in December 2018. Since the beginning we knew | ||
| + | that we had to involve Opentrons in the conversation, because we were working on a colony picking | ||
| + | extension module for the OT-2. We contacted Kristin Ellis from Opentrons and this turned out to be | ||
| + | the | ||
| + | right approach for us, because Kristin is very familiar with the OT-2 community. She has been a | ||
| + | big | ||
| + | help to us ever since by bridging us with Opentrons’ technical experts or other kinds of | ||
| + | resources. | ||
| + | At | ||
| + | the time Kristin told us that colony picking is a big topic in the OT-2 community and gave us a | ||
| + | few | ||
| + | contacts, among them: Keoni Gandall.<br> | ||
| + | <br> | ||
| + | Keoni Gandall is a bio-hacker who is determined to open source systems in Synthetic Biology. He is | ||
| + | an | ||
| + | avid user of the OT-2 because of the philosophy that OT-2 embodies: an affordable, and open-source | ||
| + | pipetting robot. Colony picking is a big part of a cloning workflow, whose automation involves a | ||
| + | lot | ||
| + | of money. There has yet to be an affordable solution for a colony picker, and Keoni Gandall | ||
| + | believes that | ||
| + | the OT-2 | ||
| + | has | ||
| + | the potential to fill this gap. When we mentioned our colony picking project to Keoni Gandall, it | ||
| + | directly | ||
| + | resonated with him, and this gave us an extra justification for our project: this is what the | ||
| + | community wants and needs. We listened to the community and let it shape our project. Since then | ||
| + | we | ||
| + | kept in touch with Keoni Gandall exchanging tips and tricks for the OT-2.<br> | ||
| + | <br> | ||
| + | </p> | ||
| + | </div> | ||
| + | </section> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div class="sub" | |
| − | + | onclick="popup('promega')"> | |
| − | + | <div class="sub-header"> | |
| − | <div class=" | + | <h1> |
| − | <h1 | + | P R O M E G A<br> |
| − | < | + | </h1> |
| + | <hr> | ||
</div> | </div> | ||
| − | <div class=" | + | <div class="sub-content"> |
| − | < | + | <div> |
| − | + | Automation of plasmid purification using the OT-2. | |
| − | + | </div> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</div> | </div> | ||
</div> | </div> | ||
| − | + | <div id="promega" | |
| − | + | class="popup"> | |
| − | + | <div class="popup-container"> | |
| − | + | <div class="popup-header"> | |
| − | + | <h1 class="title">Promega</h1> | |
| − | + | <button type="button" | |
| − | + | onclick="hide('promega')">X</button> | |
| − | + | </div> | |
| − | + | <div class="popup-content" | |
| − | + | style="text-align: justify;"> | |
| − | + | <section class="section"> | |
| − | + | <div> | |
| − | + | <figure style="float: right; margin-left: 25px;"> | |
| − | + | <img style="height: 400px; width: 600px" | |
| − | + | src="https://static.igem.org/mediawiki/2019/b/b4/T--Marburg--margarethe_schwarz.jpg" | |
| − | + | alt="Placeholder image"> | |
| − | + | <figcaption style="max-width: 600px"> | |
| − | + | Fig.1 - Margarethe Schwarz visiting the iGEM team Marburg 2019 to watch the OT-2 perform a | |
| − | + | plasmid | |
| − | + | purification using the Promegas Wizard® MagneSil® Plasmid Purification System. | |
| − | + | </figcaption> | |
| − | + | </figure> | |
| − | + | <p style="text-align: justify; margin-bottom: 1em;"> | |
| − | + | When the iGEM year started, we thought about how we could ease the work in the lab using our OT-2. | |
| − | + | We | |
| − | + | decided automating the cloning process would be a great idea and soon got into contact with | |
| − | + | Promega | |
| − | + | to | |
| − | + | tell them about our vision. Margarete Schwarz, area manager of southwest germany, and Nans Bodet, | |
| − | + | a | |
| − | + | Field Support Scientist (FSS) from the automation department at Promega, were both convinced that | |
| − | + | the | |
| − | + | automation of the cloning workflow would be a challenge, but with creativity and some work it | |
| − | + | would | |
| − | + | be | |
| − | + | a major breakthrough and a great tool for everyone with access to an OT-2.<br> | |
| − | + | <br> | |
| − | + | In a Skype call both agreed that they would love to see Promegas Wizard® MagneSil® Plasmid | |
| − | + | Purification System integrated into the workflow, being Promegas very first automated workflow in | |
| − | + | Opentrons OT-2 and the first protocol for plasmid purification in a large collection of Opentrons | |
| − | + | protocols. Promega covered our costs in terms of kits we needed for the protocols so we could | |
| − | + | focus | |
| − | + | on | |
| − | + | optimizing the workflow.<br> | |
| − | + | <br> | |
| − | + | We performed the plasmid purification a few times manually, so we would get familiar with the | |
| − | + | whole | |
| − | + | workflow and get a feeling where problems in the automated process could arise. We were in regular | |
| − | + | contact with Nans Bodet and he gave great advice on how to automate the shaking process in the | |
| − | + | OT-2 and | |
| − | + | that | |
| − | + | we would need the 8-channel pipette to scale up the number of samples that could be handled with | |
| − | + | our | |
| − | + | protocol. For the shaker he told us to get in contact with QInstruments, a company which designs | |
| − | + | and | |
| − | + | builds small shakers that are simultaneously capable of heating and cooling the samples. Thanks to | |
| − | + | recommendations from Nans Bodet a member of their support team, Ralf Paetzold, wrote us back and | |
| − | + | kindly | |
| − | + | helped us to secure a permanent loan for the BioShake D30-T elm back in June. Through a grant our | |
| − | + | team | |
| − | + | won, we were able to purchase the 8-channel pipette arm.<br> | |
| − | + | <br> | |
| − | + | When the shaker arrived, we realized it was a bit bigger than the SPS format for modules in the | |
| − | + | OT-2 | |
| − | + | and needed stabilizing support. We designed an <a | |
| − | + | href="https://2019.igem.org/Team:Marburg/Human_Practices">adapter for the shaker </a> that is | |
| − | + | robust | |
| − | + | enough to | |
| − | + | withstand the forces that occur during intense shaking.<br> | |
| − | + | <br> | |
| − | + | Furthermore, Opentrons is currently rolling out a major update from their OT-2 3.9 to 4.0 firmware | |
| − | + | that | |
| − | + | included a lot of paradigm change. This changed the way we had to define our labware and we ended | |
| − | + | up | |
| − | + | defining our shaker module coordinates as a Python dictionary importable via a JSON file. After | |
| − | + | some | |
| − | + | calibrations with our OT-2 we were trying to finish the protocol; thankfully Opentrons customer | |
| − | + | service | |
| − | + | was patient with us. They told us how to calibrate the OT-2 directly via the terminal because we | |
| − | + | had | |
| − | + | some difficulties.<br> | |
| − | + | <br> | |
| − | + | In late august Margarete Schwarz paid us a visit, curious about how the plasmid purification with | |
| − | + | Promegas Kit would perform and look like in the OT-2. We were also asked to write a <a | |
| − | + | style="padding: 0" | |
| − | + | href="https://www.promegaconnections.com/it-takes-a-village-automating-plasmid-purification-for-igem/">blog | |
| − | + | post</a> about | |
| − | + | our thoughts and progress on automating plasmid purification for the Promega Connections Blog.<br> | |
| − | + | <br> | |
| − | + | By the end of this iGEM year we were able to develop a <a style="padding: 0" | |
| − | + | href="https://2019.igem.org/Team:Marburg/Miniprep">working protocol</a> for the single-channel | |
| − | </ | + | pipette for up to 6 samples, as well as a protocol for the 8-channel pipette for up to 48 |
| − | </ | + | samples.<br> |
| − | </ | + | <br> |
| + | We are very happy about this fruit bearing interaction, we think both sides profited from this | ||
| + | cooperation in a big way.</p> | ||
| + | </div> | ||
| + | </section> | ||
| + | </div> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 1,488: | Line 1,839: | ||
</div> | </div> | ||
</div> | </div> | ||
| − | |||
</html> | </html> | ||
{{Marburg/footer}} | {{Marburg/footer}} | ||
Latest revision as of 23:46, 13 December 2019
H U M A N P R A C T I C E S

Report on Genetic Engineering
Genetic engineering has been a hotly debated topic in politics as well as society in the past decades and
still is today.
Arguments like the nutrition of a growing world population due to a declining infant mortality rate or the
loss of
considerable areas of arable land due to erosion or pollution damage keep fueling the controversy whether
genetically modified
organisms (GMO), especially crops, are needed to sustain the global demand for food. On the opposite,
concerns
have been raised
concerning the potential adverse effects on human health and environmental safety. Besides the facts, part
of
the public dispute
is based around ethical questions and trust issues towards institutions and authorities. There have been
studies and surveys
carried out addressing many of these topics. Additionally a diverse cluster of organisations and the
media
is bombarding the
public with contrary statements. This report tries to give an overview on mankind's relation towards modifying
genetics, a brief
summary of used methods, and gathers statements from scientists and authorities. It is meant as the
motivational basis for this
years Marburg iGEM team´s Public Engagement and Human Practice efforts.
G M O
R E P O R T
GMO Report
History of Genetic Modification
Our ancestors had no conception of genetics but still were able to influence the genes of multiple
organisms.
It is a
process known to everybody called artificial selection or selective breeding. Those individuals with
the
most
desirable traits,
like the biggest and most delicious fruits or the highest loyalty, is chosen to propagate and
produce
offspring. This process
is repeated over several generations and the result is an organism with the selected traits. The
dog,
existing
today in many
variations, is believed to be the organism our ancestors selectively bred first around 32,000 years
ago (Zimmer, 2013). And there are many more instances like corn which originates
from a grass called teosinte
with
very few kernels (‘Evolution of
Corn’,
n.d.). However, this process is not considered GMO technology today. What we understand under
genetic
modification today can be traced back to
the mid 1900´s,
when scientists discovered that genetic material can be transferred between different species
(Avery, MacLeod, & McCarty, 1944),
the structure of genetic material was identified as a double helix (Crick, Watson, & Bragg, 1954), the genetic code was
deciphered (Nirenberg, Matthaei,
Jones, Martin, &
Barondes, 1963) and finally a DNA recombinant technology was described (Cohen, Chang, Boyer, & Helling, 1973). Only a few
decades after these
ground-breaking discoveries were made, the first
genetically modified (GM) plants were produced in 1983, which were antibiotic resistant tobacco and
petunia
(Bevan & Chilton, 1982; Fraley,
1983; Herrera‐Estrella
et al., 1983).
Soon, the first GM plants were commercialized: In the
early 1990´s China approved modified tobacco and in 1994 the United States Food and Drug
Administration
(U.S.
FDA) approved
the “FLAVR SAVR” tomato which was modified to have a longer shelf live by delaying ripening. Today,
numerous
GM
plants exist
and are in use, covering popular fruits like papaya, melon and apple, flowers like roses, feed
plants like
sugar beet,
vegetables like tomato, maize and potato and even cotton for clothes production
(‘GM Crops
List—GM Approval
Database | ISAAA.org’, n.d.).
Current Numbers on GM Crops
World
As stated above, many GM crops are relevant for food production today, be it
indirectly
for
animal feed in
production lines or directly as consumables. In 2018, 26 countries planted 191.7 million hectares
worldwide
with GM crops,
which is an increase of 1% from 2017´s worldwide planted area. Accordingly, since its first
commercialization
in 1996 with
1.7 million hectares planted, GM crop area increased by an approximate 113-fold. The accumulated
area
planted
with GM crops
from 1996 to 2018 was 2.5 billion hectares. This makes biotechnology the fastest adopted crop
technology
in
the world. Of
the 193 member nations of the United Nations Organisation (UNO) 42 nations plus the European Union
(EU)
adopted GM crops,
of which 26 countries (21 developing and 5 industrial) planted and 44 imported GM crops. The four
major GM
crops, namely
soybeans, maize, cotton and canola, occupied 99% of the GM crop area (Figure 1). GM crop share in
total
crop
area was 78%
for soybeans, 76% for cotton, 30% for maize and 29% for canola. 42% of the global GM crop area was
planted
with
stacked trait
crops tolerant to various herbicides and pesticides. Around the world the GM crop area was unevenly
distributed with the top
five countries United States of America (USA), Brazil, Argentina, Canada and India planting 91% of
the
global
GM crop area.
In the EU, the two nations Spain and Portugal planted the GM crop MON810, which is an insecticide
resistant
maize, together
covering 120.990 hectares. 95% of the area was planted by Spain. From 2017 to 2018 GM crop area in
the EU
has
decreased by
8% from 131.535 hectares (Figure 2). Nevertheless the EU imported GM crops, roughly 30 million tons
of
soybean
products,
10 million tons of maize and 2.5 million tons of canola originating from Argentina, Brazil and the
USA.
Since
1992, across
the world 4.349 approvals to GM crops have been issued, of this being 2.063 for food, 1.461 for
animal feed use
and
825 for
cultivation
(‘ISAAA Brief 54-2018: Executive Summary | ISAAA.org’, n.d.).

*GM sugar beets, potatoes, apple, squash, papaya and brinjal/eggplant.
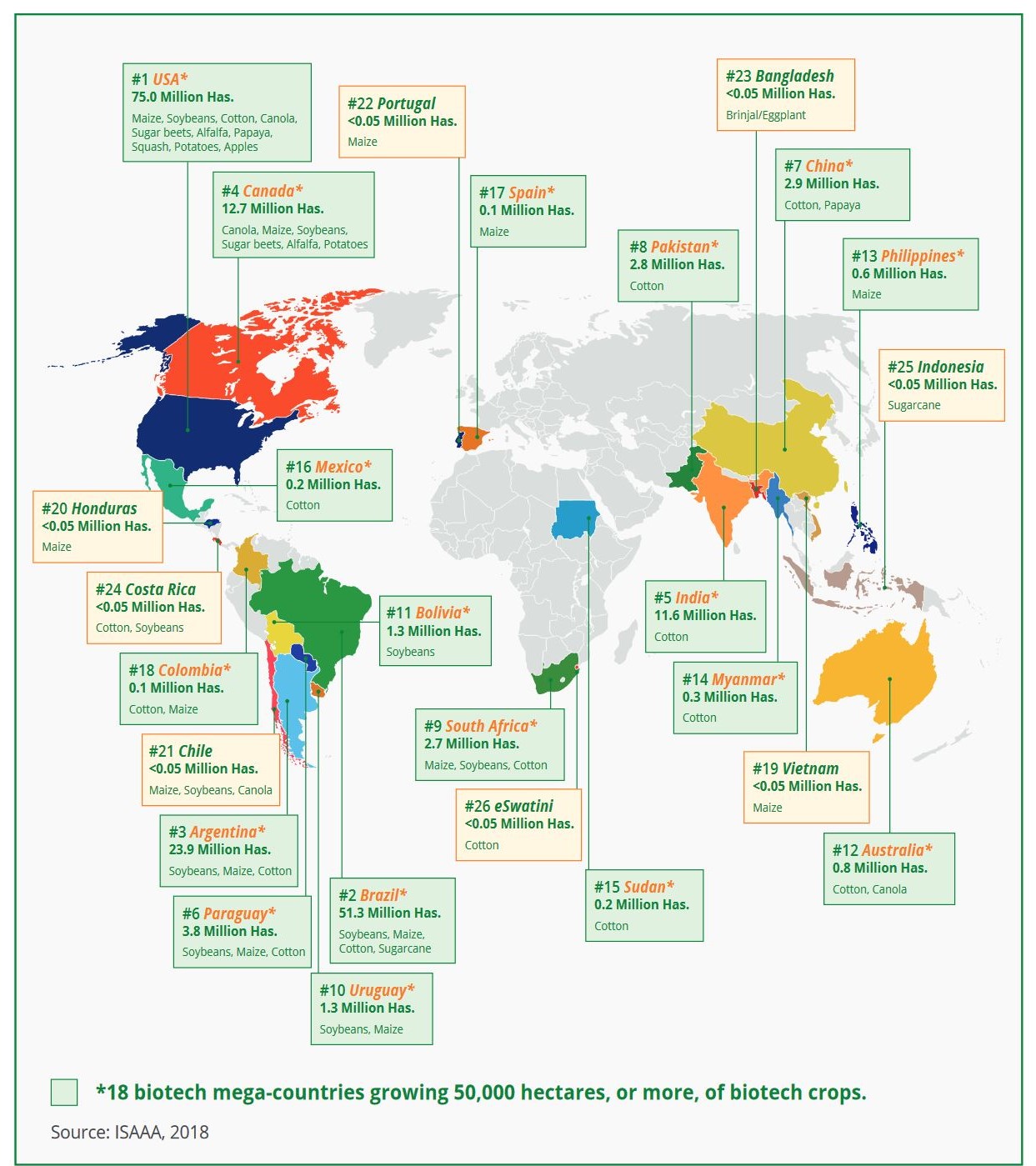
Germany
In Germany, there is no more GM crop farming since 2012. GM maize has been planted last in 2008
(3.171 hectares, 0.15% of total maize area in Germany) and GM potatoes have been planted last in
2011
(2 hectares, 0.0008% of total potato area in Germany). GM crop area never made up more than 0.02% of
land
used
by
agriculture in Germany
(‘Gentechnik’, n.d.).
Modern Methods in Breeding
The traditional way of breeding, as explained above, although having generated many domestic plants
and animals,
is
relatively
slow and limited by the available traits individuals express. Modern breeding methods enhance the
trait
spectrum and the pace
in which new traits can be discovered and implemented to crops and animals.
Plant Mutagenesis
As it is known that practical breeding depends on genetic variation plant mutagenesis expands the
variability of
traits.
Variations found in nature do not represent the original spectra of spontaneous mutations due to the
fact
that
they are
recombining within populations and interacting with environmental factors. In the process of
mutagenesis
heritable changes
occur in the genetic information induced by mutagenic agents called mutagens. These mutagens can be
of
chemical,
for
instance substances interacting with the DNA, or of physical origin, such as ionizing radiation
(Oladosu et al., 2016).
After using the mutagen on the crops, mostly seeds, seedlings or cell cultures from which single
cells can
be
grown out,
screening has to be done to see if changes in traits have been achieved by mutations. These
mutations can
be
DNA
double
strand breaks, single base exchanges or alkylation of bases. In most cases, generated mutants are
heterozygous,
because
the mutation happened in only one allele. Therefore the breeder needs to rear subsequent generations
to
evaluate
recessive
mutations. Selection then takes place in form of phenotypical, physical or molecular testing to
determine for
instance plant
height, earliness of maturity and biochemical composition. Mutagenesis breeding has impacted
agriculture
massively with
more than 3.300 entries to the Mutant Variety Database
(‘Mutant
Variety Database’,
n.d.),
covering all major food and feed crops.
Genetic Engineering
This term is used to describe methods which alter the genetic makeup of an organism using DNA
recombinant
technology.
This technology resorts to enzymatic tools called restriction enzymes. These cut the DNA site
specific and
can
thereby
isolate genetic constructs coding for desirable traits. When gene(s) are introduced into an organism
this
can be
achieved
either directly or indirectly. The direct approach utilizes a method called microparticle
bombardment
(Sanford, 1990).
Developed in the 1980´s, engineered DNA is coated on microparticles of either gold or tungsten and
then
shot with high velocity at the target organism using high pressure helium gas. The DNA fragments can
then
be
incorporated
into the organism’s genetic material. There are other direct methods such as electroporation or
microinjection
but particle
bombardment is the most effective. The indirect approach makes use of a vector: the soil bacterium
Agrobacterium
tumefaciens
naturally infects plants and alters its hosts genome via a plasmid called Ti-plasmid. This plasmid
can be
engineered to carry
genes coding for a desired trait instead of its natural genes for infection. With the development of
a
method
called
CRISPR/Cas9 and other variants genetic engineering in plants got much easier
(Cong et al., 2013;
DeMayo & Spencer, 2014;
Ran et al., 2013).
This system is found in bacteria where it serves as a defence mechanism against viruses. The
endonuclease
is
guided to its
target cutting site via a guide RNA where it induces a double strand break (DBS). The DBS can be
repaired
in
two distinct
ways. Non-homologous end joining leads to a small deletion while homologous recombination allows for
the
integration of
donor DNA into the endogenous DNA. Thereby, the CRISPR method allows for small alteration or whole
gene
insertions at target
sites.
At this point it may be appropriate to introduce the two terms “cisgenic” and
“transgenic”.
While
“transgenic”
refers to organisms in which genetic material outside the species boundary, originating from a donor
organism
which is
sexually incompatible to the engineered organism, has been inserted. “Cisgenic” on the contrary
describes
genetic
modifications within the boundaries of sexual compatibility. Therefore, cisgenic plants are similar
to
traditionally bred
plants (Schouten, Krens, & Jacobsen,
2006). The most
obvious example
of transgenic plants are the many varieties of so
called “Bt” crops. Standing for Bacillus thuringiensis, into these plants a gene from the
bacterium was
integrated which
leads to the production of a crystal protein that is toxic to specific pest insects
(‘Insecticidal
Plants’, 2015).
Opinions on GMOs
There are many scientific publications evaluating specific GMO traits towards the environment and
health
safety.
Additionally many reviews exist summarizing GMO effects to a much broader scale possible here
(Bawa & Anilakumar, 2013;
Nicolia, Manzo, Veronesi, &
Rosellini, 2014;
Snell et al., 2012;
Zhang, Wohlhueter, & Zhang,
2016).
In many of these, authors conclude that the application of GMOs offers great opportunities but still
has to
be
carried out
with precautions. A simple “yes” or “no” cannot be given
(Zhang et al., 2016). Still,
due
to the partly
contradictory
evidence, it cannot be said there is a consensus among scientists, according to
Hilbeck et al., 2015.
Benefits of GM Crops
Humanity faces several challenges in the coming decades. Amongst them are the increasing world
population, a
decrease of
arable land or the bottleneck of traditional breeding methods
(Zhang et al., 2016).
To all of these, GMOs pose a genuine answer. The easiest way to produce more food for a growing
population
is to
increase
productivity by earlier maturity, easier harvesting, processing and cultivation. Adding to that, if
we
resorted
to
organically producing todays yields, humanity would need to cultivate an additionally 3 billion
hectares,
which
is the
equivalent to the size of two South Americas
(‘Time
to call
out
the anti-GMO conspiracy theory – Mark Lynas’, n.d.).
But food also needs to become more nutritious. A good example here is “Golden Rice”
(Ye et al., 2000),
which produces a precursor of vitamin A. The deficiency of vitamin A is estimated to kill more than
half a
million
children under the age of 5 each year
(Black et al., 2008)
and cause another half a million irreversible cases of childhood blindness
(Humphrey, West, & Sommer,
1992).
Risks of GM Crops
GMOs pose risks to its consumer as do crops deriving from traditional breeding. Major risks are
toxicity,
allergenicity and genetic hazards emerging from the inserted or altered gene itself, the expressed
protein,
products
of the metabolism, pleiotropic effects or the disruption of natural genes in the organism
(Zhang et al., 2016).
There have been reports on the strong allergenicity of “Starlink” maize, which is directly connected
to
the
inserted gene
from Bacillus thuringiensis
(Bravo, Gill, & Soberón,
2007;
Sanchis, 2011;
Tabashnik, 1994;
Werth, Boucher, Thornby, Walker, & Charles,
2013).
Also, GM crops can have an adverse ecological influence. For example, the weed species Amaranthus
palmeri
did
evolve a
glyphosate resistance after years of glyphosate use on resistant cotton fields
(Gilbert, 2013).
Another possibility is the fact, that insect resistant crops infer with ecological food webs by
shifting
predator prey
ratios. Moreover, targeted pests might decline and primary minor pests become major issues
(Bawa & Anilakumar, 2013; Snow &
Palma, 1997).
Statements from Authorities
The "Public Acceptance of Agricultural Biotechnologies (PABE) project" revealed a range of questions
concerning
rather institutional considerations of the public, such as who is benifitting from GMO use, by whom
consequences
have
been evaluated, if authorities have enough power to regulate large companies and why the public has
not
been
better
informed about GMO usage
(Marris, 2001).
For this reason, an overview of institutional statements might be appropriate.
The European Commision (EC) published the book “A decade of EU-funded GMO research”. Within this
endeavor
more
than
200 million Euro of research grants were spent to evaluate GMO´s in areas such as environmental
impact,
food
safety,
biomaterials, biofuels, risk assessment and management. It conclusively states: “The main conclusion
to be
drawn from the efforts of more than 130 research projects, covering a period of more than 25 years
of
research,
and
involving more than 500 independent research groups, is that biotechnology, and in particular GMOs,
are
not
per
se more
risky than e.g. conventional plant breeding technologies.”
(Publications
Office of the European Union, 2010).
The National Academy of Sciences founded by the U.S. Congress summarizes in their comprehensive
report,
that
large
numbers of animal feeding studies provided reasonable evidence that animals were not harmed by food
derived
from
GM crops,
although admitting some studies were not designed optimal. Furthermore, long-term data in livestock
health
before and
after GM crop introduction did not show adverse effects associated with the crops. And at last,
epidemiological
data on
cancer and human health over time was revised but no substantiated evidence was found that GM crops
are
less
safe than
foods from non-GM crops
(Read "Genetically Engineered Crops,
n.d.).
The British Royal Society states the following to the question “Is it safe to eat GM crops?” on its
website:
“Yes.
There is no evidence that a crop is dangerous to eat just because it is GM. There could be risks
associated
with
the
specific new gene introduced, which is why each crop with a new characteristic introduced by GM is
subject
to
close scrutiny.
Since the first widespread commercialisation of GM produce 18 years ago there has been no evidence
of ill
effects linked to
the consumption of any approved GM crop.” Before new GM foods are permitted to the market a variety
of
test
has
to be
completed and the results are used by the authorities to determine the safety of the GM product,
making
“new
GM
crop
varieties at least as safe to eat as new non GM varieties, which are not tested in this way.”
(‘Is it
safe
to
eat GM crops?’, n.d.).
Conclusion
As biologists, using genetic engineering methods every single day, they are quite natural to us.
Nevertheless,
we are
confronted with the public debate too. Having experienced the public aversion towards GMO ourselves
and
having
read about
the many proposed justifications against it we realized that a direct exchange between the public
and
experts
from all
fields as well as diverse interest groups might provide a good common ground for an open discussion.
D R . N I N A
S C H E E R
Dr. Nina Scheer
Burning forests, melting ice. With greenhouse gases as a global threat to our climate we need a
multitude of ways to solve this issue. One of the biggest challenges in the future will be
nutrition: the UN expects the world population to rise to 10 billion by 2050. Without genetically
engineered crops it will be very hard to nourish such a huge population.
We met Nina Scheer from the Social Democratic Party of Germany (SPD) one month before our panel
discussion to bring politics and science closer together. In particular, we discussed green genetic
engineering. Like most of germany’s political parties Nina Scheer and the SPD are in suspicion about
genetic engineering. A general fear for possible consequences is holding innovation at stagnation,
preventing any further development and leaving important key technologies to global competitors.
As we discussed with Dr. Scheer about our project and green genetical engineering in general, she
supported the idea that
the state should invest more funding into research, rather than us relying on third-party funds.
Nonetheless, Dr. Scheer did see a trust issue concerning irreversible damages to nature and humankind
as
well as a lack of good control mechanisms when using green genetic engineering. At the example of
Contergan® Nina Scheer highlighted the challenges of uncertainty. We learned that we as scientists
have
the duty to proof the unmitigated safety of our products and beyond that not only proof it to our
own community but also to bring this trust to society as a whole if we want our research to have an
impact on reality.
We thank Dr. Nina Scheer for this opportunity to discuss major issues of our time.
P L A N T M A R K E T
Plant Market

We advertised our panel discussion by running a booth at the plant breeders festival in Marburg on the weekend of 14th September. It is an event for young and old people alike and the perfect platform to bring our discussion closer to a diverse audience.
When we talked with Nina Scheer, member of the German parliament from the Social Democratic Party, she told us that a large part of the population has a very critical attitude towards genetically modified food. We are convinced that the main reason for this critical attitude is a lack of information and communication between scientists and the public that leads to insecurities in the population. In order to get a picture of the public opinion on genetic engineering, we asked the participants to fill out a questionnaire, which we then statistically evaluated. We realized that in particular middle-aged and older people have a critical view on genetic engineering or tend to say from themselves that they don't know enough about it and would like to hear more. Through personal discussions we were able to get in contact with people, raise awareness on the issue and also learned a lot ourselves about the landscape of opinions.
At the same time we gave children and interested adults the chance to take a closer look at our work by letting them experimentally extract DNA from pepper (figure 1). We used this to also explain to the children what DNA is, what it means to them and what exactly they were doing in each step.
Due to the contact and discussion with people from the general society and especially outside of our university background, the plant breeder festival expanded our knowledge and we therefore see it as a great success. We received a lot of positive feedback and spoke personally with many supporters and critics of genetic engineering and took away some lively discussions. In addition, we were able to evaluate around 200 questionnaires and further promote our panel discussion. This was demonstrated by the run on our subsequent panel discussion, where we were happy to recognize some familiar faces from the festival.
Evaluation
In our questionnaire, we first had the participants assess their current state of knowledge about
genetic engineering and divided the questions into gender, age and basic attitude.
As far as gender is of interest, most of the participants rated themselves in the criteria as "rather bad" and "medium". Men consider themselves to be better informed than women. Especially in the category "very good" it is mainly men, while in the category "very bad" it is mostly women, regarding their self-assessment. It is possible that men are more self-confident in this aspect and thus, consider themselves to be better informed.

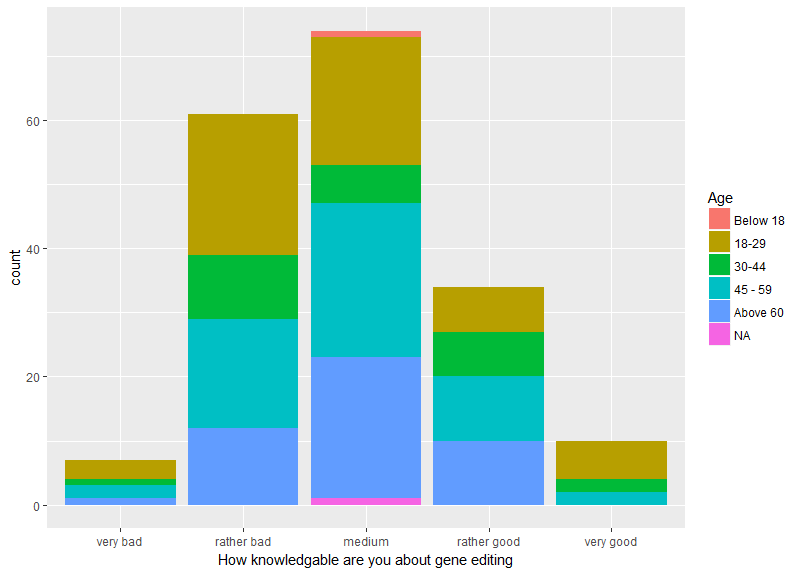

In the category “age”, the average self-assessment is also "rather bad" and "medium". However, younger people (18-29) generally considered themselves to be better informed than older people. A problem is that the older generation may have more difficulty accessing information.
It is also noticeable that the participants have a positive attitude towards genetic engineering when they are better informed. This shows that there is not enough information on genetic engineering and that it does not reach the population.
The first part of our study reveals two problems: bad accessibility of information to the older generation creates a discrepancy between the level of knowledge of younger and older people. As a result, older people feel less informed, which in turn leads to skepticism towards genetic engineering. For example, one can focus the source of information on television/radio or newspaper and thus try to reach older people more effectively.There is also a problem with the flow of information in general, as people are obviously better attuned to genetic engineering when they know more about it. In general, there is a need for better information on the subject and more discussions at a political level.
We then asked whether the participants considered genetic engineering in plants, animals and humans to be ethically justifiable.
Most people had a positive attitude towards plants. Here, many people probably recognized the potential, also with regard to the 2050 food problem or resistance to pests.
Surprisingly, many participants are more likely to agree with genetic engineering in
humans than in animals. Most people probably think of the potential of genetic engineering to fight
serious diseases and thus cure diseases where traditional treatment is not sufficient. In addition,
when
it comes to genetic engineering on animals, some people may think of animal experiments, which many
consider to be reprehensible. In addition, it is potentially possible to ask people before such
interventions, while animals have no voice. Our results confirmed us conducting our panel discussion
to
create another source of information and to inspire as many people as possible to think about it. We
hope to be able to educate and hope that the potential of genetic engineering will be recognized by
the
general public.



P A N E L D I S C U S S I O N
Genetic engineering panel discussion report: Blessing or curse
Genetic engineering has been a hotly debated topic in politics as well as in society in the past
decades and still is today. New methods in Genome Editing (GE) are making the regulation of methods
more and more important due to the increased difficulty to identify changes in the genetic material
of manipulated organisms. The European Court of Justice (ECJ) ruling on GE of July 2018 pleased the
interest groups of genetic engineering sceptics, but disappointed the research community to a large
extent. The panel discussion organised by the iGEM team Marburg 2019 with participants from plant
breeding research, industry, ethics research and genetic engineering critics should give the public
an insight into the different breeding methods and the according viewpoints of different
stakeholders. After a keynote lecture by biochemist Prof. Dr. Lars-Oliver Essen from the Philipps-
University of Marburg the panel discussion started. It was moderated by Michael Lange of "Forschung
aktuell" from Deutschlandfunk and team member Paula Mueller. The energetic audience, in which all
age groups were represented, soon actively took part in the discussion, asked questions and
criticized statements of the discussants on stage.
In his keynote lecture, Prof. Dr. Lars-Oliver Essen tried to convey the scientific context of the
evening to the audience. Using various common plants, such as bananas and carrots, he illustrated
the products of classical breeding methods through human selection. All the species mentioned above
originate from primitive types, which differ from today's food plants, for example by an enormously
lower proportion of fruit flesh. He then outlined the differences between more modern breeding
methods such as chemical mutagenesis or radiation and genetic tools such as the headline-making
CRISPR genetic scissors. At the end of his keynote lecture, Prof. Dr. Essen outlined the ECJ ruling
of July 2018 and gave an outlook on topics that influence the genetic engineering discussion, such
as the population growth of mankind and the patenting of modern genetic engineering methods as well
as the resulting products.

Most of the panelists agreed that genetic engineering offers opportunities, especially in
agriculture, to address challenges such as climate change or the nutrition of the growing world
population. According to the introductory statement of Prof. Dr. Andreas Weber from the Cluster of
Excellence on Plant
Science at Heinrich-Heine University in Düsseldorf, methods of genetic
engineering such as CRISPR are already established tools and have no cause for concern, particularly
in fundamental research. But subsequently, modified organisms cannot be tested in the field under
natural
conditions. However, the objective of testing and checking modified plants is a problem, because
often independent testing organisations do not have access to certified seeds and the complete
data used in the designing process, replied Dr. Christoph Then from testbiotech e.V.
It was also discussed whether the product or the manufacturing process should serve as the basis for
evaluation. Since a plant modified by GE methods cannot be distinguished from a plant cultivated by
established breeding methods, the product should be the basis of the review, argued Daniel
Stukenberg, representative of the party "The Humanists". A point of view with a drastically
different background, here represented by ethics professor Dr. Friedemann Voigt, put the consumers in
the foreground and demanded their right to know how a food is produced. This led the discussion
further in the direction of the effects of genetically modified animal feed on farm animals and the
human
consumer. At this point, Dr. Christoph Then criticised the sharp decline in risk research, which
pursues society's protective goals with regard to health.
The panel also separated into simple gene edited and transgenic organisms. While gene edited plants
do not contain genes from foreign organisms, such genes in transgenic plants can, for example,
provide certain ingredients that make them resistant to pests or insecticides/herbicides. Transgenic
plants must be assessed with special attention and in each unique case, since the individual
composition of the ingredients is specific for each modified organism. This point also met with the
approval of the audience, which was expressed by strong applause. Transgenic plants could not
develop in a natural way and should therefore not be used in agriculture. At the moment, however,
the use of genetically modified plants in organic farming is also not in compliance with
regulations, according to Freya Schäfer of FiBL (Research Institute of Organic Agriculture).

Paula Müller, Prof. Dr. Friedemann Voigt, Dr. Claus Kremoser, Daniel Stukenberg, Freya Schäfer, Dr. Christoph Then,
Prof. Dr. Andreas Weber and Michael Lange.
Prof. Dr. Friedemann Voigt continued to ensure the ethical part of the discussion. He noted that
compared to the rest of the world, there is a certain mistrust towards genetic engineering,
especially in Europe. One had to estimate the risks against the opportunities, whereby he mainly
identified an acceptance problem in the population, which could not be met scientifically. However,
there is not only distrust of genetic engineering, but also of the economic motivation to use it.
Dr. Claus Kremoser expressed concerns about seed companies with patent monopolies that could promote
large monocultures. Nevertheless, the development and application of genetic engineering is strongly
dependent on the respective motivation: patents are the driving force in the industry while in the
scientific community it is the pressure to publicise. Too strict and thus very costly requirements
for the testing of genetically modified organisms make it impossible for small companies to
participate in the competition. Consequently, strong regulation would lead to a few large
monopolies.
The discussion ended with the consensus that genetic engineering in agriculture should be regulated
responsibly and objectively. However, due to ideological blockades and diffuse fears in society and
among political influencers, a meaningful willingness to compromise was very low.
Integrated Human Practices
C Y A N O
B I O T E C H
Cyano Biotech
Cyano Biotech is one of the leading, top edge companies to work with sustainable and
product-oriented
phototrophic bacteria in Germany. As a shining figure in cyanobacterial metabolic engineering, the
CEO
of Cyano Biotech Dr. Dan Enke talked with us about the possibilities of our project. His feedback
led
us to the design of adjusting specific parameters in our growth experiments and to the integration of
our
terminator library. Through our talk with a potential end user of our Green
Expansion
and our
engineered
strains, we achieved a more “real-world” focus of our project.
As our project was in its beginning, one thing was clear: high throughput methods are essential for
any
viable chassis in Synthetic Biology. Sadly, we quickly noticed obstacles in our way, such as
inhibited
growth in our plates compared to flasks. In consequence of that, we looked for help and who would be
better, than an expert on that specific field? We talked to Dan Enke and he kindly provided us with
data in his own well plate experiments. Soon we noticed, that he inoculated cultures several times
in a
row out of the exponential phase, leading to a huge boost in doubling times. Through this support we
were able to design and conduct a growth curve with as much as five precultures.
Aside from that, we talked about another facet of our project, cyanobacterial terminators. Being
overread in
cyano´s, both metaphorically and for real, they are one of the most important
regulatory genetic elements. Yet another expert in the
field of cyanobacteria was pleased of our idea to set on the search for the best terminator and
strongly encouraged us to utilize the Green Expansion to its full extent.

P R O F. D R.
A N N E G R E T W I L D E
Cultivation expertise from leading cyano scientist Prof. Wilde
While working with the cyanobacterium Synechococcus elongatus UTEX 2973 we noticed very
quickly a
lack of
standardization in the field of Synthetic Biology. To tackle this huge problem, we
decided to focus, as one of our main goals, on standardization to make scientific results more
comparable. Therefore, we worked on standardizing light measurement, cultivating parameters
(temperature, CO2, rpm, …) and the cultivation media for cyanobacteria, especially UTEX 2973.

During the theoretical planning of our project we contacted scientists in research and industry
that
are specialists for cyanobacteria. As a result, Prof. Dr. Annegret Wilde (Institute of Biology,
Albert-Ludwigs-University Freiburg) answered our call and was very interested in advising us in
regards to our projects. In order to give us an introduction to the handling of cyanobacteria, she
invited us to her institute at the University of Freiburg on 5th and 6th June 2019. In a short and
focused internship we were able to quickly gain a set of core competencies regarding sterile
inoculation, streaking of cyano cultures and further information regarding the cultivation
conditions
which we applied to our strain.
In addition, we also learned that the measurement of light intensity is an important topic. There
is
a
variety of measuring instruments and different methods for each, which means that information on
light
intensities should be viewed with caution. For cyanobacteria such as Synechococcus
elongatus UTEX
2973
light intensity plays a decisive role, which is why we analysed differences between a variety of
instruments and methods to establish a standard for light measurement based on our results .
In further discussions about our Marburg Collection 2.0 we were recommended to take a very close
look
at terminators as they oddly enough have an effect on the transcription of
upstream
genes. As a
result, we decided to take a closer look at that and investigated their effects. We thank
Professor
A.
Wilde for her input, her invitation to Freiburg and her recommendations that guided us in our
project.
D O U L I X
Doulix
“My major bottleneck is colony picking.” - Davide De Lucrezia, Managing Director of Doulix
The vision of automating colony picking has existed now for many years. Big companies like Tecan,
Singer Instruments or Hudson Robotics invented robots those are able to identify and pick
colonies.
The problem is that they cost a fortune, starting at 50,000€ up to 100,000€ or even more. Although
it is highly desired, a low cost solution is still missing for this tedious task. Start-ups like
Doulix are currently automating such workflows in their laboratories, but they do not have the
funds
to finance a state-of-the-art colony picking robot. By now every single step has to be performed
manually, draining resources from other departments, which actually should be paid more attention
to. “Colony picking is a bottleneck in every of part our workflows” says Davide De Lucrezia, the
founder and managing director of Doulix. Doulix focuses on developing innovative technologies for
scientists to simplify their work, especially in the field of Synthetic Biology, and they are
currently planning on establishing Opentrons OT-2 in their lab to automate the most part of their
workflows.
During an online conference with Davide De Lucrezia, Sota Hirano, and Alessandro Filisetti from
Doulix, Davide De Lucrezia suggested that turning the OT-2 into a colony picker as a project would
be
really
interesting. To have a fully trained, ready to use package to turn the OT-2 into a colony picker
would enhance the workflow at Doulix tremendously. Nevertheless, to suit the user's needs as well
as
to get this job done in the spirit of Opentrons, installing the needed add-ons should be as
modular
and flexible as possible and designed so that “even a biologist” without technical knowledge or
programming skills would be able to install and use them.
That is where our team came into play. We decided to take this advice to our heart and started to
work out what was needed to turn the OT-2 into a fully automated colony picking robot.

One of the first big design questions was whether we wanted to hardcode an image recognition
software for the colony detection or if it was a better choice to train a data hungry but - given
proper and enough training data - more accurate and scalable artificificial intelligence based
colony detection. Kristin Ellis, the director of strategic initiatives at Opentrons referred us to
Keoni Gandall from Stanford, a well known tinker of the OT-2 for more unconventional
applications.
He is building a colony picking system himself, however he chose not to rely on an AI. He
recommended us to go with AI as he thinks his approach is very prone to changes in parameters. If
many
different users want to utilize the same system, a flexible software is required that can take
environmental changes into account. We decided to opt for maximum flexibility by working with an
AI.
Now that we had an idea of the required software we started to design modular hardware to overcome
potential problems in a fully automated workflow in the OT-2. To illuminate the agar plates in the
right
way without any distortions we
engineered a light table that distributes light equally over the plate.
To give an “eyesight” to the OT-2 we mounted a Raspberry Pi 4 and an ArduCAM on the OT-2 arm. For
a
better accessibility we created our Graphical User Interface for Directed Engineering (GUIDE). We
designed our GUI in a way that will enable every user to train their own AI with their own
training
data set so that the AI can be optimized for each specific situation. Moreover the GUI will also
enable access to users that are not trained in computer/ software engineering.
Now that we gave our robot the ability to see, to think and to communicate with us, nothing stood
in
the way of our own colony picker. We are now able to turn the OT-2 into a colony picker costing
below $300. Moreover, for companies, teams or
groups who do not own an OT-2 yet, we were able to reduce the costs for purchasing a colony picker
by 90-95%, compared to the listed market prices for traditional colony pickers.
Finally we contacted Doulix the second time to discuss in more details about our project. They
approved it and gave further suggestions such as “live training” so that the AI will continue to
learn as it is being used to pick up colonies. Not only will this improve the AI gradually but it
will also adapt to the specific needs of each user. This leaves a lot of room to improve the
project
in the future.
S T A N D A R D I Z A T I O N
Standardization in Cyanocommunity

During our visit at the Cyano Conference 2019 in Tübingen we recognized a need for standardization
in
this community. We asked people to send us their BG-11 recipes and
surprisingly
received several different versions. The community at the conference was aware of the missing
standardization and we received very positive feedback for our efforts. Fixed standards are
essential
for reproducibility of results, especially the preparation of media and buffers but no one is
investing time to set a standard. After the cyano conference we stayed in contact with Nicolas
Schmelling, coordinator of the bachelor program at CEPLAS. During his PhD he was working on
establishing more standards in the cyano community. He tried to establish protocols and collected
different methods and recipes to establish a standard for all.
After he clarified to us the importance of comparable media in context of standards, we started to
collect different BG11 recipes and compared them in growing experiments. Figure 1
depicts our results and shows the impact of different recipes for media.
In our experiment we could show that there is a significant difference between the different BG11
recipes despite their relative similarity. During our complete project we were working on the
standardization for light intensity, media and different cultivating parameters. We made it our
destiny
to make the first step into the beginning of
standardization in
the cyano community by providing Synechococcus elongatus with standardized
parameters. We
were able to find the optimal growing conditions for UTEX 2973 and could show that creating a
standard
in measurement and methods is really important to have comparable results. With our project we
hope
that we could set a first step into standardization, so that the future cyano community will have
standardized and comparable results.
C Y A N O
C O N F E R E N C E 2 0 1 9
The CYANO Conference 2019 in Tübingen
From September 11th to September 13th we attended the CYANO Conference 2019 in Tuebingen funded by the VAAM (Vereinigung für Allgemeine und Angewandte Mikrobiologie). During the poster sessions we took the chance to present our project and how we revolutionize the upcoming work on phototrophic organisms. Therefore, we gained great feedback from the participants, which showed huge interest in our toolbox specified for cyanobacteria. Our Synthetic Biology approaches encountered the thinking of classical cyanobacterial research which lead to interesting discussions from which we gained a lot of input. Furthermore, the leading experts of cyanobacteria offered talks where we learned how to modify working on Synechococcus elongatus.
We were especially interested in the discussions about methods. We soon realized that the cyanobacterial scene has no standardized protocols for daily laboratory practices and they are also aware of that issue. This started with debates about the media composition of BG-11 media but also concerned issues like standardized evaluation of light conditions. With our project for standardizing growth conditions and providing a part collection we tackle these major issues for scientists studying phototrophic organisms.

E X P E R T O N C Y A N O S :
J A M E S G O L D E N
Expert on Cyanos - James Golden
While diving deeper and deeper into the ocean of possibilities that cyanobacteria have to offer we
noticed a few inconsistencies in literature.
BG11 media is commonly used in cyanobacterial research, but the exact composition seemed to be
different across every second paper we read. Optical densities are more frequently measured at a
wavelength of 730nm, though 750nm seems to be the better choice. For Synechococcus
elongatus
UTEX 2973
the “optimal growth conditions” according to literature are often quite different; some state 38°C
and
500µE at a CO2 level of 3% fits best, others prefer 41°C and 1500µE with 5% CO2 concentration.
But how are these light intensities measured? With a planar device or a
spherical
one? We have not
seen this being explained in literature.
As all of these things have an incredibly huge impact on various different experiments we saw the
need
to find a standardized answer to our questions, reaching out to as many experts in this field as
we
could reach - whether it was industry or research.
One of the leading laboratories working with cyanobacteria is the Golden Lab of the UC San Diego.
Susan Golden and her husband James W. Golden have both been working with cyanobacteria for quite
some
time, now with a stronger focus on their use for industrial purposes.
We set up a Skype call with them, but sadly Susan Golden was not able to join us on short
notice.
During our talk with James W. Golden we laid open our concerns about the cyanobacterial community
and
he quickly supported our train of thoughts, as he himself noticed a lack of standardization. He
assured us that this is a hot topic in this field of research, as many do not seem to care enough
about the reproducibility of their data and encouraged us to continue with our efforts.
More accurately, he talked with us about why he thinks there is still no clear decision on whether
to
measure optical densities at 730nm or 750nm: It might be a technical problem, as many photometers
are
simply not able to measure wavelengths of 750nm. In contrast, he mentioned that 750nm would be the
more optimal way, as it proves to minimize absorbance from pigments in cyanobacterial cells,
presenting more accurate data. This confronted us with a conflicting decision: Would it be better
to
use the more accurate 750nm or 730nm, as the latter would allow more labs all over the world to
measure in the same way.
This was one of the key factors that led us to measure the whole spectrum of our cultures for our
growth curves, as this would provide a larger dataset, awarding us not just with 730nm and 750nm
data,
but also the possibility to check if the spectrum shows normal behavior, from which one could
conclude
how healthy the cultures are.

In the beginning we measured the light intensity of our incubators with a planar
measurement device - the only one available for us. Talking to James Golden we realized that we
should
try to get hold of a spherical measurement device, as he assured us that this is the way to
generate
more accurate data, leading to a more reproducible setup - exactly what we were aiming for.
After receiving such a device from Biospherical Instruments, we again implemented the feedback we
got and
measured growth curves.
One with cultures at 1500µE measured with a spherical device and one with 1500µE measured with a
planar device, where the measured intensities were converted to theoretically spherical values
with
a
conversion chart offered to us by Prof. Dr. Annegret Wilde from the University of Freiburg.
These experiments were a huge step in our project, as they heavily influenced the way we cultured
our
cyanobacteria, not only drastically improving their growth, but also clearly demonstrating how
flawed
certain measurements can be.
We would never have been able to reach the fast doubling times we achieve now without this crucial
input and as this will be the case for others too, we made it our mission to keep on stressing the
importance of this way of measurement whenever we reach out to the scientific community.
Again, thank you very much Prof. Dr. James W. Golden for your invaluable contribution!
E X P E R T O N C Y A N O S:
N I C O L A S S C H M E L L I N G
Expert on Cyanos - Nicolas Schmelling
The composition of BG11 media is another important issue we were able to discuss with Nicolas
Schmelling.
While working in our own lab we already got the notion that not all BG11 media are prepared in the
same
way, which is the reason why we kindly asked other researchers from the cyano community- like
James Golden
- to send us their recipes. In order to compare the various ways the BG11 media can be prepared,
we tried
those recipes and measured growth curves to find the perfect fit.


It was clear that the growth of our cultures was comparably fast at the beginning no matter what
media
was used, but one of them stood out: BGM - it enabled faster growth at higher ODs, allowing
cultures
to reach double the OD of other cultures after the same time.
We are certain that having the same ideal medium throughout different cyano labs is not just
elemental
for optimal growth, but also vital for comparability, as trying to reproduce the growth conditions
of
papers can be quite tricky when it is not clear what exact medium was used and how it was
prepared.

O P E N T R O N S
& K E O N I G A N D A L L
Opentrons + Keoni Gandall
“A colony picking module for the OT-2 will be a great help” - Keoni Gandall
We started with the colony picking project back in December 2018. Since the beginning we knew
that we had to involve Opentrons in the conversation, because we were working on a colony picking
extension module for the OT-2. We contacted Kristin Ellis from Opentrons and this turned out to be
the
right approach for us, because Kristin is very familiar with the OT-2 community. She has been a
big
help to us ever since by bridging us with Opentrons’ technical experts or other kinds of
resources.
At
the time Kristin told us that colony picking is a big topic in the OT-2 community and gave us a
few
contacts, among them: Keoni Gandall.
Keoni Gandall is a bio-hacker who is determined to open source systems in Synthetic Biology. He is
an
avid user of the OT-2 because of the philosophy that OT-2 embodies: an affordable, and open-source
pipetting robot. Colony picking is a big part of a cloning workflow, whose automation involves a
lot
of money. There has yet to be an affordable solution for a colony picker, and Keoni Gandall
believes that
the OT-2
has
the potential to fill this gap. When we mentioned our colony picking project to Keoni Gandall, it
directly
resonated with him, and this gave us an extra justification for our project: this is what the
community wants and needs. We listened to the community and let it shape our project. Since then
we
kept in touch with Keoni Gandall exchanging tips and tricks for the OT-2.
P R O M E G A
Promega

When the iGEM year started, we thought about how we could ease the work in the lab using our OT-2.
We
decided automating the cloning process would be a great idea and soon got into contact with
Promega
to
tell them about our vision. Margarete Schwarz, area manager of southwest germany, and Nans Bodet,
a
Field Support Scientist (FSS) from the automation department at Promega, were both convinced that
the
automation of the cloning workflow would be a challenge, but with creativity and some work it
would
be
a major breakthrough and a great tool for everyone with access to an OT-2.
In a Skype call both agreed that they would love to see Promegas Wizard® MagneSil® Plasmid
Purification System integrated into the workflow, being Promegas very first automated workflow in
Opentrons OT-2 and the first protocol for plasmid purification in a large collection of Opentrons
protocols. Promega covered our costs in terms of kits we needed for the protocols so we could
focus
on
optimizing the workflow.
We performed the plasmid purification a few times manually, so we would get familiar with the
whole
workflow and get a feeling where problems in the automated process could arise. We were in regular
contact with Nans Bodet and he gave great advice on how to automate the shaking process in the
OT-2 and
that
we would need the 8-channel pipette to scale up the number of samples that could be handled with
our
protocol. For the shaker he told us to get in contact with QInstruments, a company which designs
and
builds small shakers that are simultaneously capable of heating and cooling the samples. Thanks to
recommendations from Nans Bodet a member of their support team, Ralf Paetzold, wrote us back and
kindly
helped us to secure a permanent loan for the BioShake D30-T elm back in June. Through a grant our
team
won, we were able to purchase the 8-channel pipette arm.
When the shaker arrived, we realized it was a bit bigger than the SPS format for modules in the
OT-2
and needed stabilizing support. We designed an adapter for the shaker that is
robust
enough to
withstand the forces that occur during intense shaking.
Furthermore, Opentrons is currently rolling out a major update from their OT-2 3.9 to 4.0 firmware
that
included a lot of paradigm change. This changed the way we had to define our labware and we ended
up
defining our shaker module coordinates as a Python dictionary importable via a JSON file. After
some
calibrations with our OT-2 we were trying to finish the protocol; thankfully Opentrons customer
service
was patient with us. They told us how to calibrate the OT-2 directly via the terminal because we
had
some difficulties.
In late august Margarete Schwarz paid us a visit, curious about how the plasmid purification with
Promegas Kit would perform and look like in the OT-2. We were also asked to write a blog
post about
our thoughts and progress on automating plasmid purification for the Promega Connections Blog.
By the end of this iGEM year we were able to develop a working protocol for the single-channel
pipette for up to 6 samples, as well as a protocol for the 8-channel pipette for up to 48
samples.
We are very happy about this fruit bearing interaction, we think both sides profited from this
cooperation in a big way.
